7XXC
 
 | | Orf1-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
3B5A
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating A38G mutation with a 2'OMe modification at the active site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B5F
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating the Ade38Dap Mutation and a 2',5' Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3ZOW
 
 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3BBM
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38C and 2'O-Me Modification at Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBK
 
 | | Miminally Junctioned Hairpin Ribozyme Incorporates A38C and 2'5'-phosphodiester Linkage within Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
7YFC
 
 | | Cryo-EM structure of the histamine-bound histamine H4 receptor and Gq complex | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YFD
 
 | | Cryo-EM structure of the imetit-bound histamine H4 receptor and Gq complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, CHOLESTEROL, Engineered G-alpha-q, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
7Z8Q
 
 | | Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Brodolin, K. | | Deposit date: | 2022-03-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Nat Commun, 14, 2023
|
|
9BTS
 
 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
3B5S
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates A38DAP Mutation and 2'-O-methyl Modification at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
8XIE
 
 | | Archaeal exosome complex (Rrp4-Rrp41-Rrp42) | | Descriptor: | Exosome complex component Rrp4, Exosome complex component Rrp41, Exosome complex component Rrp42 | | Authors: | Kim, H.S, Park, S.H, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Insights into the Rrp4 Subunit from the Crystal Structure of the Thermoplasma acidophilum Exosome.
Biomolecules, 14, 2024
|
|
4AGA
 
 | | Hofmeister effects of ionic liquids in protein crystallization: direct and water-mediated interactions | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Mukhopadhyay, A, Carvalho, A.L, Romao, M.J. | | Deposit date: | 2012-01-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hofmeister Effects of Ionic Liquids in Protein Crystallization: Direct and Water-Mediated Interactions
Cryst.Eng.Comm., 14, 2012
|
|
7XQ9
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
2PFC
 
 | | Structure of Mycobacterium tuberculosis Rv0098 | | Descriptor: | Hypothetical protein Rv0098/MT0107, PALMITIC ACID | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence.
Chem.Biol., 14, 2007
|
|
8W0U
 
 | | Human LCAD complexed with Acetoacetyl Coenzyme A | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, Long-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8W0T
 
 | | Human LCAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8W0Z
 
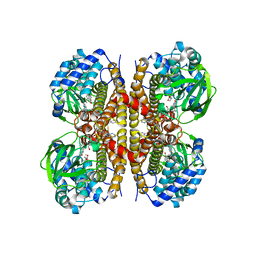 | | Human LCAD complexed with Lauric Acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LAURIC ACID, Long-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
7YPU
 
 | | OrfE-CoA-glycylthricin complex | | Descriptor: | Acetyltransferase, COENZYME A, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7YPV
 
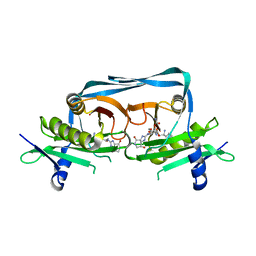 | | Crystal structure of OrE-ST-F | | Descriptor: | Acetyltransferase, Streptothricin F | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
3B91
 
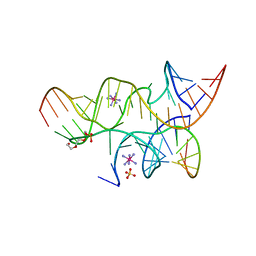 | | Minimally Hinged Hairpin Ribozyme Incorporates Ade38(2AP) and 2',5'-Phosphodiester Linkage Mutations at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
2Q8D
 
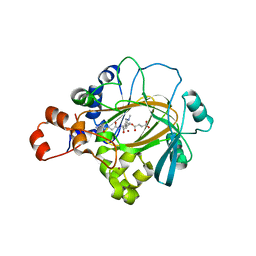 | | Crystal structure of JMJ2D2A in ternary complex with histone H3-K36me2 and succinate | | Descriptor: | HISTONE 3 peptide, JmjC domain-containing histone demethylation protein 3A, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q8E
 
 | | Specificity and Mechanism of JMJD2A, a Trimethyllysine-Specific Histone Demethylase | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3B58
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
