1H0M
 
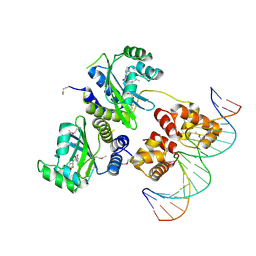 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
1BHC
 
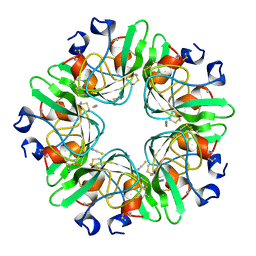 | |
2AKQ
 
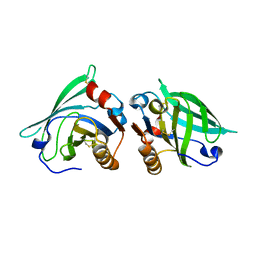 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
1GTK
 
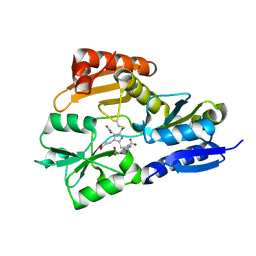 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1H0Y
 
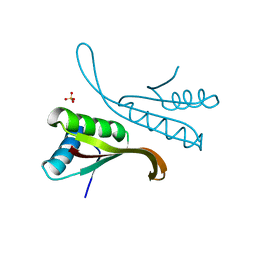 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B, SULFATE ION | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
3LQS
 
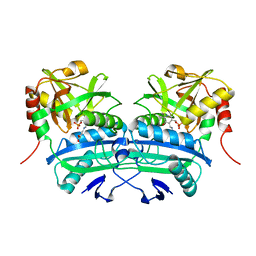 | | Complex Structure of D-Amino Acid Aminotransferase and 4-amino-4,5-dihydro-thiophenecarboxylic acid (ADTA) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, ACETIC ACID, D-alanine aminotransferase | | Authors: | Lepore, B.W, Liu, D, Peng, Y, Fu, M, Yasuda, C, Manning, J.M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chiral discrimination among aminotransferases: inactivation by 4-amino-4,5-dihydrothiophenecarboxylic acid.
Biochemistry, 49, 2010
|
|
12E8
 
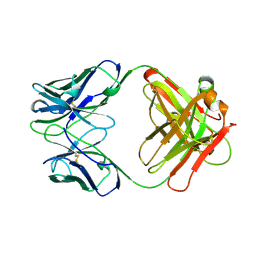 | | 2E8 FAB FRAGMENT | | Descriptor: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | Authors: | Rupp, B, Trakhanov, S. | | Deposit date: | 1998-03-14 | | Release date: | 1998-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
2B0F
 
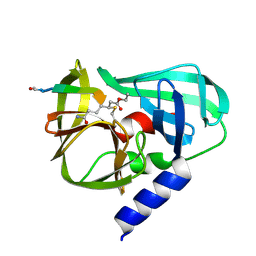 | |
3FYY
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
6ISD
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with sulcotrione | | Descriptor: | 2-[2-chloro-4-(methylsulfonyl)benzoyl]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
3MPC
 
 | | The crystal structure of a Fn3-like protein from Clostridium thermocellum | | Descriptor: | Fn3-like protein, SULFATE ION | | Authors: | Alahuhta, M.P, Xu, Q, Brunecky, R, Lunin, V.V. | | Deposit date: | 2010-04-26 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a fibronectin type III-like module from Clostridium thermocellum.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3GC8
 
 | | The structure of p38beta C162S in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, NICKEL (II) ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1AM5
 
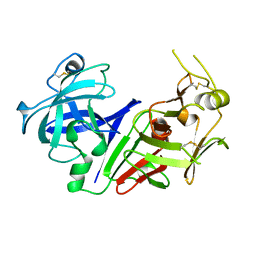 | |
3LQE
 
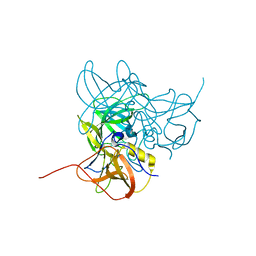 | |
1H32
 
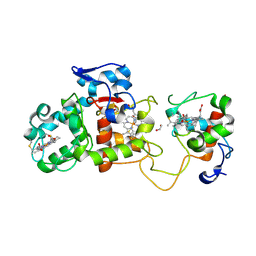 | | Reduced SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME C, DIHEME CYTOCHROME C, ... | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
7EL2
 
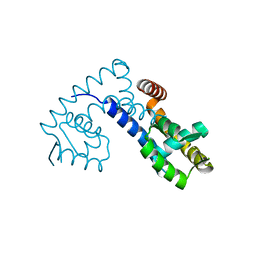 | |
7EL3
 
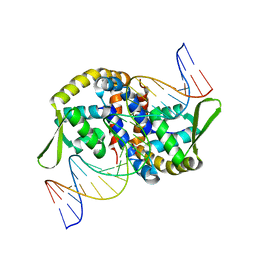 | |
1H33
 
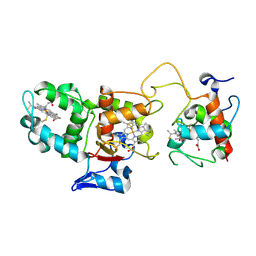 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
1H31
 
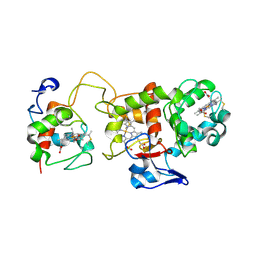 | | Oxidised SoxAX complex from Rhodovulum sulfidophilum | | Descriptor: | CYTOCHROME C, DIHEME CYTOCHROME C, HEME C | | Authors: | Bamford, V.A, Bruno, S, Rasmussen, T, Appia-Ayme, C, Cheesman, M.R, Berks, B.C, Hemmings, A.M. | | Deposit date: | 2002-08-21 | | Release date: | 2002-11-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Oxidation of Thiosulfate by a Sulfur Cycle Enzyme
Embo J., 21, 2002
|
|
3GC7
 
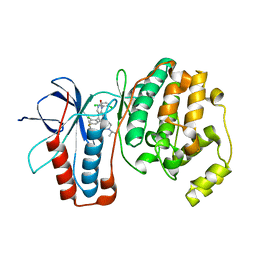 | | The structure of p38alpha in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6J63
 
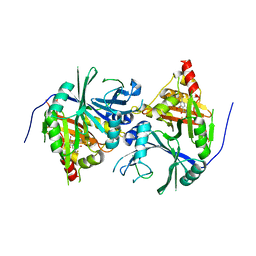 | | Crystal structure of Arabidopsis thaliana HPPD complexed with NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2019-01-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
6JDP
 
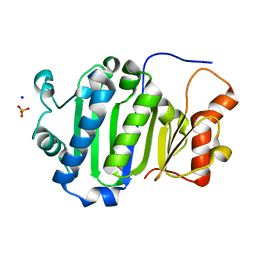 | |
1GK7
 
 | | HUMAN VIMENTIN COIL 1A FRAGMENT (1A) | | Descriptor: | SULFATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
5V85
 
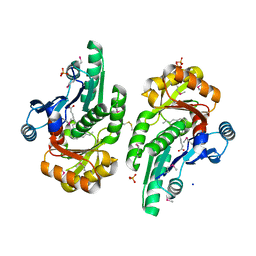 | | The crystal structure of the protein of DegV family COG1307 from Ruminococcus gnavus ATCC 29149 (alternative refinement of PDB 3JR7 with Vaccenic acid) | | Descriptor: | EDD domain protein, DegV family, PHOSPHATE ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
5OFT
 
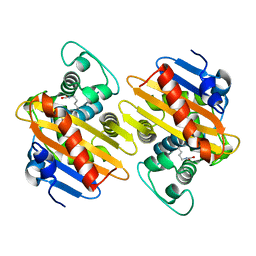 | |
