7Q8V
 
 | | Crystal structure of TTBK1 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 1, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q8W
 
 | | Crystal structure of TTBK1 in complex with VNG1.35 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Tau-tubulin kinase 1, ... | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7QV1
 
 | | Bacillus subtilis collided disome (Leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QV2
 
 | | Bacillus subtilis collided disome (Collided 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QH4
 
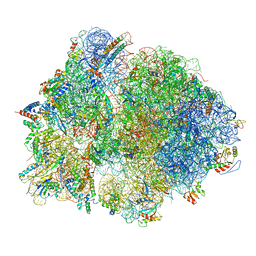 | | Structure of the B. subtilis disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (5.45 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QV3
 
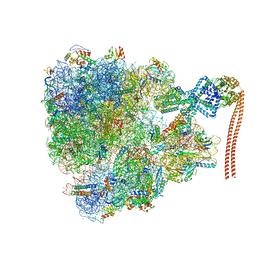 | |
7QGU
 
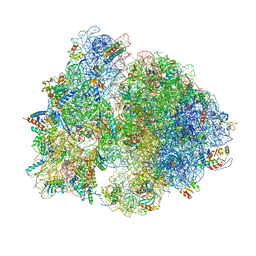 | | Structure of the B. subtilis disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
3L44
 
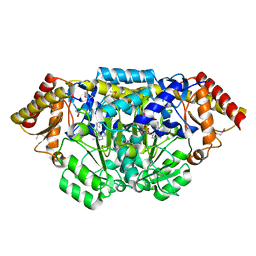 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
7QHW
 
 | | TTBK1 kinase domain in complex with inhibitor 29 | | Descriptor: | GLYCEROL, SULFATE ION, Tau-tubulin kinase 1, ... | | Authors: | Nozal, V, Liehta, D. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
8BUU
 
 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-11-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
3J2Q
 
 | | Model of membrane-bound factor VIII organized in 2D crystals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Stoilova-Mcphie, S, Lynch, G.C, Ludtke, S, Pettitt, B.M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (15 Å) | | Cite: | Domain organization of membrane-bound factor VIII.
Biopolymers, 99, 2013
|
|
5OA1
 
 | | RNA polymerase I pre-initiation complex | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Sadian, Y, Tafur, L, Kosinski, J, Jakobi, A.J, Muller, C.W. | | Deposit date: | 2017-06-20 | | Release date: | 2017-07-26 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into transcription initiation by yeast RNA polymerase I.
EMBO J., 36, 2017
|
|
3J2S
 
 | |
8BW4
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW2
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-N-(2-methoxyethyl)-2-methyl-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
4NFN
 
 | | Human tau tubulin kinase 1 (TTBK1) complexed with 3-({5-[(4-amino-4-methylpiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-bromophenol | | Descriptor: | 3-({5-[(4-amino-4-methylpiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-bromophenol, SULFATE ION, Tau-tubulin kinase 1 | | Authors: | Sheriff, S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of human tau-tubulin kinase 1 both in the apo form and in complex with an inhibitor.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
6D2Q
 
 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
4N90
 
 | |
4NFM
 
 | | Human tau tubulin kinase 1 (TTBK1) | | Descriptor: | GLYCEROL, Tau-tubulin kinase 1 | | Authors: | Sheriff, S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of human tau-tubulin kinase 1 both in the apo form and in complex with an inhibitor.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5RJP
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024672 | | Descriptor: | 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK3
 
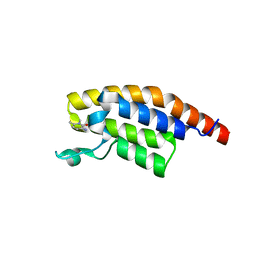 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1501469697 | | Descriptor: | 3-amino-1,6-dimethylpyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKN
 
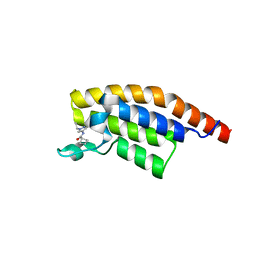 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z373768900 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJW
 
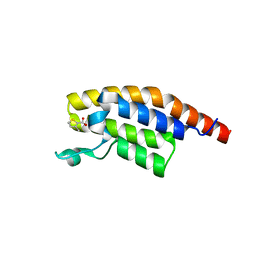 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z256709556 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKB
 
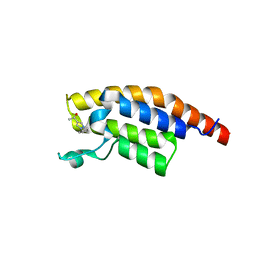 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z2004563941 | | Descriptor: | (1S)-1-(1-cyclopentyl-1H-pyrazol-4-yl)ethan-1-ol, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
