6L0C
 
 | |
6CX6
 
 | |
4R0V
 
 | | [FeFe]-hydrogenase Oxygen Inactivation is Initiated by the Modification and Degradation of the H cluster 2Fe Subcluster | | Descriptor: | ARSENIC, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Swanson, S.D, Ratzloff, M.W, Mulder, D.W, Artz, J.H, Ghose, S, Hoffman, A, White, S, Zadvornyy, O.A, Broderick, J.B, Bothner, B, King, P.W, Peters, J.W. | | Deposit date: | 2014-08-01 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster.
J.Am.Chem.Soc., 137, 2015
|
|
4GNJ
 
 | | Crystal Structure Analysis of Leishmania siamensis Triosephosphate Isomerase | | Descriptor: | ARSENIC, SODIUM ION, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Leartsakulpanich, U. | | Deposit date: | 2012-08-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of Leishmania siamensis Triosephosphate Isomerase
To be Published
|
|
3H6N
 
 | | Crystal Structure of the ubiquitin-like domain of plexin D1 | | Descriptor: | ARSENIC, Plexin-D1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Nedyalkova, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Buck, M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal Structure of the ubiquitin-like domain of plexin D1
To be Published
|
|
6IG5
 
 | |
4CYS
 
 | | G6 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa, in complex with Phenylphosphonic acid | | Descriptor: | AMMONIUM ION, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6IEN
 
 | | Substrate/product bound Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ARGININOSUCCINATE, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
6UDN
 
 | | Crystal Structure of a Self-Assembling DNA Scaffold Containing TA Sticky Ends and Rhombohedral Symmetry | | Descriptor: | ARSENIC, DNA (5'-D(*TP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6IEM
 
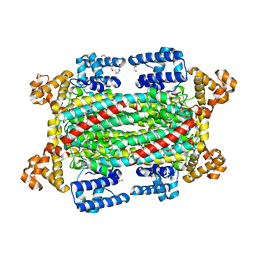 | | Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, Argininosuccinate lyase, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
4CXK
 
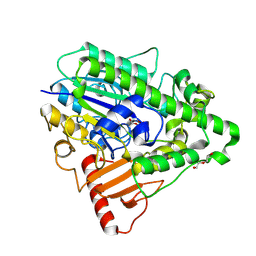 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6J05
 
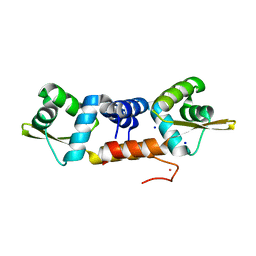 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
4CYR
 
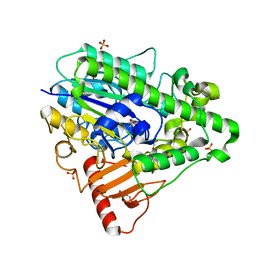 | | G4 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2E9F
 
 | |
1HDH
 
 | | Arylsulfatase from Pseudomonas aeruginosa | | Descriptor: | Arylsulfatase, CALCIUM ION, SULFATE ION | | Authors: | Boltes, I, Czapinska, H, Kahnert, A, von Buelow, R, Dirks, T, Schmidt, B, von Figura, K, Kertesz, M.A, Uson, I. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 A Structure of Arylsulfatase from Pseudomonas Aeruginosa Establishes the Catalytic Mechanism of Sulfate Ester Cleavage in the Sulfatase Family.
Structure, 9, 2001
|
|
6IGA
 
 | |
9GGI
 
 | | Crystal structure of argininosuccinate lyase from Arabidopsis thaliana (AtASL) | | Descriptor: | Argininosuccinate lyase, chloroplastic, CHLORIDE ION, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A.J, Krzeszewska, D, Sekula, B. | | Deposit date: | 2024-08-13 | | Release date: | 2024-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Arabidopsis thaliana argininosuccinate lyase structure uncovers the role of serine as the catalytic base.
J.Struct.Biol., 216, 2024
|
|
9GGJ
 
 | | Crystal structure of argininosuccinate lyase from Arabidopsis thaliana (AtASL) in complex with biological substrate and products - argininosuccinate, argnine and fumarate | | Descriptor: | ARGININE, ARGININOSUCCINATE, Argininosuccinate lyase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A.J, Krzeszewska, D, Sekula, B. | | Deposit date: | 2024-08-13 | | Release date: | 2024-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Arabidopsis thaliana argininosuccinate lyase structure uncovers the role of serine as the catalytic base.
J.Struct.Biol., 216, 2024
|
|
2Y0O
 
 | |
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
1AOS
 
 | |
5JH0
 
 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.18 Angstrom resolution | | Descriptor: | ARS-binding factor 2, mitochondrial, DNA (5'-D(*AP*AP*TP*AP*AP*TP*AP*AP*AP*TP*TP*AP*TP*AP*TP*AP*AP*TP*AP*TP*AP*A)-3'), ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
1K62
 
 | |
4FSD
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with As(III) | | Descriptor: | ARSENIC, Arsenic methyltransferase, CALCIUM ION, ... | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
3JRN
 
 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
