8FP6
 
 | |
6WIO
 
 | | Fab antigen complex | | Descriptor: | Fab Heavy chain, Fab Light chain, Interleukin-17A | | Authors: | Antonysamy, S. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
8IC6
 
 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8FMF
 
 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6WEY
 
 | |
8I30
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | Descriptor: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
8FPM
 
 | |
8FSF
 
 | |
5JIO
 
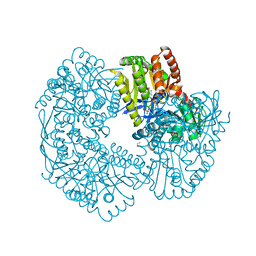 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase ternary complex with ADP and Glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
8FT2
 
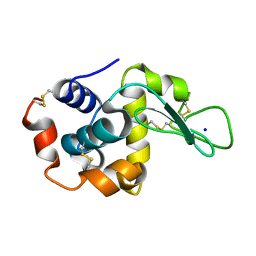 | |
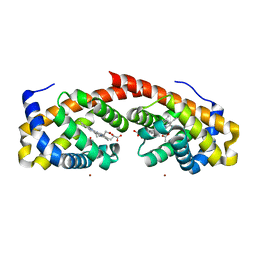
6WK3
 
 | |
8FV3
 
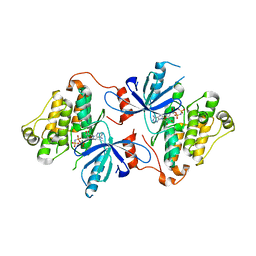 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
5JNF
 
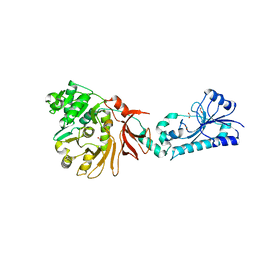 | |
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8FYY
 
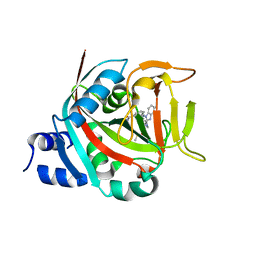 | |
5JGI
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with M45 compound | | Descriptor: | N-ALPHA-L-ACETYL-ARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
8FM1
 
 | |
6WMV
 
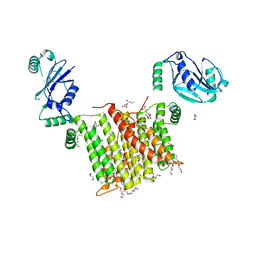 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,3',3''-phosphanetriyltripropanoic acid, AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F. | | Deposit date: | 2020-04-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
5JIQ
 
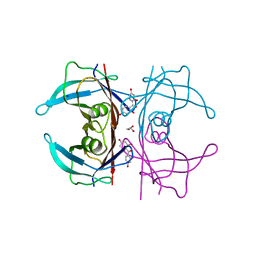 | | Crystal Structure of Human Transthyretin in Complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | Descriptor: | GLYCEROL, SODIUM ION, Transthyretin, ... | | Authors: | Begum, A, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-04-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
8FMG
 
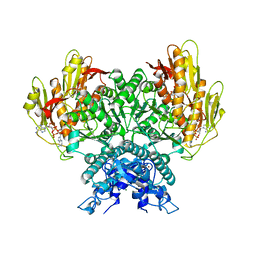 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (3 tetramers in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, MAGNESIUM ION, SAVED domain-containing protein, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMH
 
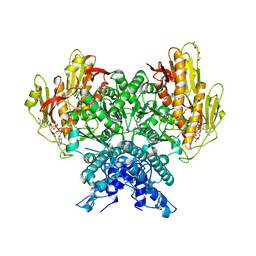 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5JIB
 
 | |
5B0E
 
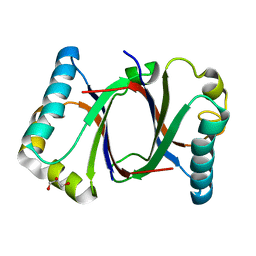 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8HZZ
 
 | | GuApiGT (UGT79B74) | | Descriptor: | SULFATE ION, apiosyltransferase | | Authors: | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
