2YNY
 
 | | Salmonella enterica SadA 255-302 fused to GCN4 adaptors (SadAK1) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Albrecht, R, Lupas, A.N. | | Deposit date: | 2012-10-20 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Complete Fiber Structures of Complex Trimeric Autotransporter Adhesins Conserved in Enterobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TIF
 
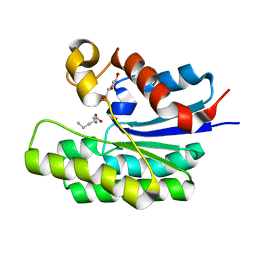 | | x-ray structure of acyl-CoA thioesterase I, TesA, triple mutant M141L/Y145K/L146K in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, DI(HYDROXYETHYL)ETHER, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
6PXU
 
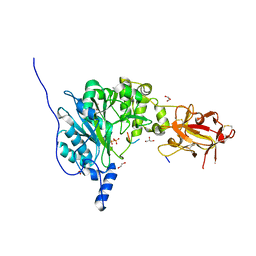 | | Crystal structure of human GalNAc-T12 bound to a diglycosylated peptide, Mn2+, and UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GAGATGAGAGYYITPRTGAGA, ... | | Authors: | Samara, N.L, Fernandez, A.J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | The structure of the colorectal cancer-associated enzyme GalNAc-T12 reveals how nonconserved residues dictate its function.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XT2
 
 | | EQADH-NADH-HEPTAFLUOROBUTANOL, P21 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,2,3,3,4,4,4-heptafluorobutan-1-ol, ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2020-07-16 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative binding modes in abortive NADH-alcohol complexes of horse liver alcohol dehydrogenase.
Arch.Biochem.Biophys., 701, 2021
|
|
1H1C
 
 | |
3REZ
 
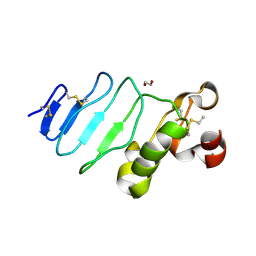 | | glycoprotein GPIb variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-05 | | Release date: | 2012-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIb beta and a GPIb beta/GPIX chimera
Blood, 118, 2011
|
|
5NWE
 
 | | Complex of H275Y mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZ4
 
 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
3QN6
 
 | |
5NZF
 
 | | Complex of H275Y/I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZN
 
 | | Complex of H275Y/S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2017-05-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZE
 
 | | Complex of S247N mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Hejdanek, J. | | Deposit date: | 2017-05-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5TIE
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 7.5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
3RFE
 
 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
3QPG
 
 | |
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
5ZST
 
 | |
5E0J
 
 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (21-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(12~{S},15~{S},18~{S})-15-(cyclohexylmethyl)-12-(hydroxymethyl)-9,14,17-tris(oxidanylidene)-1,8,13,16,21,22-hexazabicyclo[18.2.1]tricosa-20(23),21-dien-18-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
6HAZ
 
 | | Crystal structure of the bromodomain of human SMARCA2 in complex with SMARCA-BD ligand | | Descriptor: | 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Steurer, S, Weiss-Puxbaum, A, Zoephel, A, Roy, M, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
4XOJ
 
 | | Structure of bovine trypsin in complex with analogues of sunflower inhibitor 1 (SFTI-1) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Golik, P, Malicki, S, Grudnik, P, Karna, N, Debowski, D, Legowska, A, Wladyka, B, Gitlin, A, Brzozowski, K, Dubin, G, Rolka, K. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Investigation of Serine-Proteinase-Catalyzed Peptide Splicing in Analogues of Sunflower Trypsin Inhibitor 1 (SFTI-1).
Chembiochem, 16, 2015
|
|
3MYW
 
 | | The Bowman-Birk type inhibitor from mung bean in ternary complex with porcine trypsin | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Engh, R.A, Bode, W, Huber, R, Lin, G, Chi, C. | | Deposit date: | 2010-05-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin.
Eur.J.Biochem., 212, 1993
|
|
5ZSK
 
 | |
5ZSP
 
 | | NifS from Hydrogenimonas thermophila | | Descriptor: | Cysteine desulfurase | | Authors: | Nakamura, R, Fujishiro, T, Takahashi, Y. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
5ZSO
 
 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Fujishiro, T, Takahashi, Y. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6U85
 
 | |
