2YWK
 
 | | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11 | | Descriptor: | Putative RNA-binding protein 11 | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Uchikubo-Kamo, T, Nishino, A, Morita, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11
To be Published
|
|
2YY8
 
 | |
2YTZ
 
 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Homocystein in the orthorhombic crystal-lattice | | Descriptor: | N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
2YZ7
 
 | | X-ray analyses of 3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase | | Authors: | Hoque, M.M, Juan, E.C.M, Shimizu, S, Hossain, M.T, Takenaka, A. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structures of Alcaligenes faecalisD-3-hydroxybutyrate dehydrogenase before and after NAD(+) and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2Z0P
 
 | | Crystal structure of PH domain of Bruton's tyrosine kinase | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Tyrosine-protein kinase BTK, ZINC ION | | Authors: | Murayama, K, Kato-Murayama, M, Mishima, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of the Bruton's tyrosine kinase PH domain with phosphatidylinositol
Biochem.Biophys.Res.Commun., 377, 2008
|
|
2Z1C
 
 | | Crystal structure of HypC from Thermococcus kodakaraensis KOD1 | | Descriptor: | GLYCEROL, Hydrogenase expression/formation protein HypC, TETRAETHYLENE GLYCOL | | Authors: | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2007-05-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
4ITF
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(1H-tetrazole-1-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-1-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
2YQC
 
 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YVH
 
 | |
4KP0
 
 | | Crystal Structure of the human Chymase with TJK002 | | Descriptor: | 4-({1-[(4-methyl-1-benzothiophen-3-yl)methyl]-1H-benzimidazol-2-yl}sulfanyl)butanoic acid, Chymase | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-05-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a complex of human chymase with its benzimidazole derived inhibitor
J.SYNCHROTRON RADIAT., 20, 2013
|
|
2YY0
 
 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | Descriptor: | C-Myc-binding protein | | Authors: | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
2YZC
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with allantoate | | Descriptor: | ALLANTOATE ION, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
4L26
 
 | | Crystal structure of DNA duplex containing consecutive T-T mispairs (Br-derivative) | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*TP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Kondo, J, Yamada, T, Hirose, C, Tanaka, Y, Ono, A. | | Deposit date: | 2013-06-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo DNA Duplex Containing Consecutive Watson-Crick-like T-Hg(II) -T Base Pairs
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4J7A
 
 | | Crystal Structure of Est25 - a Bacterial Homolog of Hormone-Sensitive Lipase from a Metagenomic Library | | Descriptor: | Esterase | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Park, K.S, Joo, S.B, Kim, K.K, Kim, D.H. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library
Acta Crystallogr.,Sect.D, 69, 2014
|
|
4HZX
 
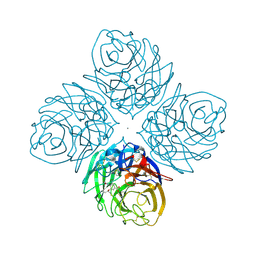 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4L71
 
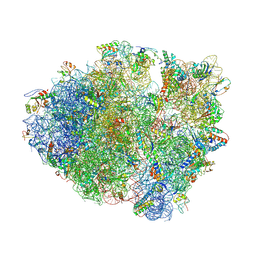 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.900001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KXF
 
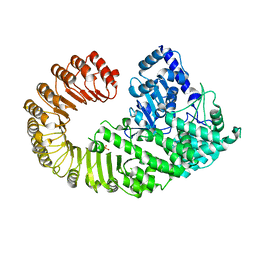 | |
4L47
 
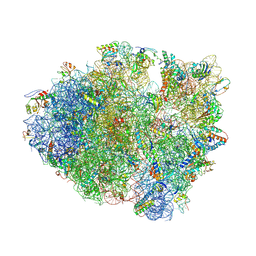 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.220001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LFZ
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.92000055 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6PEP
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgIJ from the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Protein InvG, Protein PrgH, Protein PrgI, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
8WV1
 
 | |
8X5T
 
 | | Crystal structure of Thermus thermophilus peptidyl-tRNA hydrolase in complex with adenosine 5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Uehara, Y, Matsumoto, A, Nakazawa, T, Fukuta, A, Ando, K, Oka, N, Uchiumi, T, Ito, K. | | Deposit date: | 2023-11-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode between peptidyl-tRNA hydrolase and the peptidyl-A76 moiety of the substrate.
J.Biol.Chem., 301, 2025
|
|
8WZS
 
 | |
8X0O
 
 | |
8XKP
 
 | | Crystal structure of human tyrosine-protein kinase Fes/Fps in complex with compound 17c | | Descriptor: | 7-[[(1R,2S)-2-azanylcyclohexyl]amino]-5-[[3-[(2S,6R)-2,6-dimethylmorpholin-4-yl]phenyl]amino]-4-oxidanylidene-3H-pyrido[3,4-d]pyridazine-8-carbonitrile, SULFATE ION, Tyrosine-protein kinase Fes/Fps | | Authors: | Baba, D, Hanzawa, H. | | Deposit date: | 2023-12-23 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Novel Pyrido-pyridazinone Derivatives as FER Tyrosine Kinase Inhibitors, Leading to the Potent DS08701581.
Acs Med.Chem.Lett., 15, 2024
|
|
