2BSI
 
 | |
6R5D
 
 | |
6R4U
 
 | |
6R5E
 
 | |
7ZHS
 
 | | 3D reconstruction of the cylindrical assembly of DnaJA2 delta G/F by imposing D5 symmetry | | Descriptor: | Ubiquitin-like protein SMT3,DnaJ homolog subfamily A member 2, ZINC ION | | Authors: | Cuellar, J, Velasco-Carneros, L, Santiago, C, Martin-Benito, J, Valpuesta, J, Muga, A. | | Deposit date: | 2022-04-07 | | Release date: | 2023-07-26 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The self-association equilibrium of DNAJA2 regulates its interaction with unfolded substrate proteins and with Hsc70.
Nat Commun, 14, 2023
|
|
7ZVE
 
 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
4KH6
 
 | |
8C4H
 
 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly multimer | | Descriptor: | Nucleocapsid, RNA (84-MER) | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-04 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
8B0H
 
 | | 2C9, C5b9-CD59 cryoEM structure | | Descriptor: | CD59 glycoprotein, Complement C5, Complement component C6, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8CEV
 
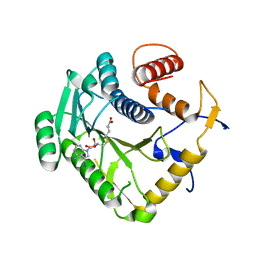 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO1119 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-iodanyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CER
 
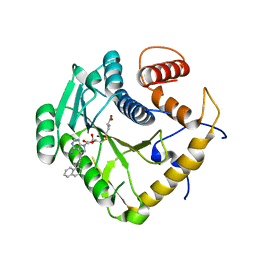 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO494 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-naphthalen-1-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CEQ
 
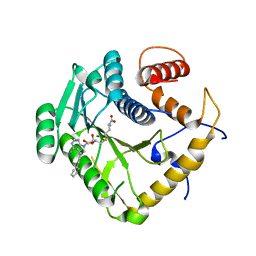 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO427 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-phenylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CES
 
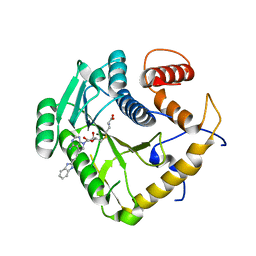 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO500 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-[2-(1~{H}-benzimidazol-2-yl)ethynyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CET
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO507 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
2B52
 
 | | Human cyclin dependent kinase 2 (CDK2) complexed with DPH-042562 | | Descriptor: | 1-(3-(2,4-DIMETHYLTHIAZOL-5-YL)-4-OXO-2,4-DIHYDROINDENO[1,2-C]PYRAZOL-5-YL)-3-(4-METHYLPIPERAZIN-1-YL)UREA, Cell division protein kinase 2 | | Authors: | Muckelbauer, J. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Synthesis and Evaluation of Indenopyrazoles as Cyclin-Dependent Kinase Inhibitors. Part 4: Heterocycles at C3
Bioorg.Med.Chem.Lett., 14, 2004
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8AWI
 
 | | Crystal structure of Human Transthyretin at 1.15 Angstrom resolution | | Descriptor: | SODIUM ION, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
5E1S
 
 | | The Crystal structure of INSR Tyrosine Kinase in complex with the Inhibitor BI 885578 | | Descriptor: | (5R)-N-(1-{2-[4-(2-methoxyethyl)piperazin-1-yl]ethyl}-1H-pyrazol-3-yl)-5,8-dimethyl-9-phenyl-6,8-dihydro-5H-pyrazolo[3,4-h]quinazolin-2-amine, Insulin receptor | | Authors: | Kessler, D, Zahn, S, Sanderson, M, Wolkerstorfer, B. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | BI 885578, a Novel IGF1R/INSR Tyrosine Kinase Inhibitor with Pharmacokinetic Properties That Dissociate Antitumor Efficacy and Perturbation of Glucose Homeostasis.
Mol.Cancer Ther., 14, 2015
|
|
2ANE
 
 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
4KH4
 
 | | Toxoplasma gondii NTPDase1 C258S/C268S in complex with Mg and AMPPNP | | Descriptor: | MAGNESIUM ION, Nucleoside-triphosphatase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Krug, U, Totzauer, R, Strater, N. | | Deposit date: | 2013-04-30 | | Release date: | 2013-11-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The ATP/ADP substrate specificity switch between Toxoplasma gondii NTPDase1 and NTPDase3 is caused by an altered mode of binding of the substrate base.
Chembiochem, 14, 2013
|
|
4KH5
 
 | | Toxoplasma gondii NTPDase1 C258S/C268S in complex with Mg and AMPNP | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, MAGNESIUM ION, Nucleoside-triphosphatase 2 | | Authors: | Krug, U, Totzauer, R, Strater, N. | | Deposit date: | 2013-04-30 | | Release date: | 2013-11-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The ATP/ADP substrate specificity switch between Toxoplasma gondii NTPDase1 and NTPDase3 is caused by an altered mode of binding of the substrate base.
Chembiochem, 14, 2013
|
|
2H6J
 
 | | Crystal Structure of the Beta F145A Rhodococcus Proteasome | | Descriptor: | Proteasome alpha-type subunit 1, Proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Proteasome assembly triggers a switch required for active-site maturation.
Structure, 14, 2006
|
|
2CME
 
 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
3GJS
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Ac-YVAD-Cho inhibitor, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
1XBO
 
 | | PTP1B complexed with Isoxazole Carboxylic Acid | | Descriptor: | 5-(3-{3-[3-HYDROXY-2-(METHOXYCARBONYL)PHENOXY]PROPENYL}PHENYL)-4-(HYDROXYMETHYL)ISOXAZOLE-3-CARBOXYLIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Zhao, H, Liu, G, Xin, Z, Serby, M, Pei, Z, Szczepankiewicz, B.G, Hajduk, P.J, Abad-Zapatero, C, Hutchins, C.W, Lubben, T.H, Ballaron, S.J, Hassach, D.L, Kaszubska, W, Rondinone, C.M, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoxazole carboxylic acids as protein tyrosine phosphatase 1B (PTP1B) inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
