1WQ2
 
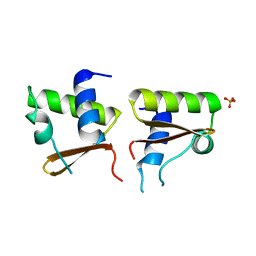 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1QJ5
 
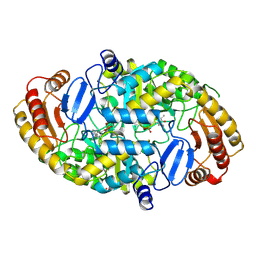 | | Crystal structure of 7,8-diaminopelargonic acid synthase | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kack, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
1J7I
 
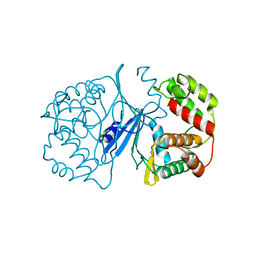 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1GSQ
 
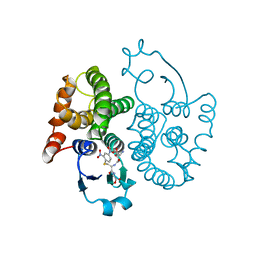 | | THREE-DIMENSIONAL STRUCTURE, CATALYTIC PROPERTIES AND EVOLUTION OF A SIGMA CLASS GLUTATHIONE TRANSFERASE FROM SQUID, A PROGENITOR OF THE LENS-CRYSTALLINS OF CEPHALOPODS | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Ji, X, Rosenvinge, E.C.V, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1995-01-09 | | Release date: | 1995-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods.
Biochemistry, 34, 1995
|
|
1PWB
 
 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D with maltose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-glucopyranose, ... | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
1PW9
 
 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
1ZAK
 
 | |
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
1WBA
 
 | | WINGED BEAN ALBUMIN 1 | | Descriptor: | WINGED BEAN ALBUMIN 1 | | Authors: | Mccoy, A.J. | | Deposit date: | 1996-06-19 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallization and preliminary crystallographic data of the major albumin from Psophocarpus tetragonolobus (L.) DC.
J.Biol.Chem., 262, 1987
|
|
1CJM
 
 | | HUMAN SULT1A3 WITH SULFATE BOUND | | Descriptor: | PROTEIN (ARYL SULFOTRANSFERASE), SULFATE ION | | Authors: | Bidwell, L.M, Mcmanus, M.E, Gaedigk, A, Kakuta, Y, Negishi, M, Pedersen, L, Martin, J.L. | | Deposit date: | 1999-04-18 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human catecholamine sulfotransferase.
J.Mol.Biol., 293, 1999
|
|
1LEH
 
 | | LEUCINE DEHYDROGENASE FROM BACILLUS SPHAERICUS | | Descriptor: | LEUCINE DEHYDROGENASE | | Authors: | Baker, P.J, Turnbull, A.P, Sedelnikova, S.E, Stillman, T.J, Rice, D.W. | | Deposit date: | 1995-06-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A role for quaternary structure in the substrate specificity of leucine dehydrogenase.
Structure, 3, 1995
|
|
1BN5
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-07-31 | | Release date: | 1999-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1BTU
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4R)-1-TOLUENESULPHONYL-3-ETHYL-AZETIDIN-2-ONE-4-CARBOXYLIC ACID | | Descriptor: | (3R)-3-ethyl-N-[(4-methylphenyl)sulfonyl]-L-aspartic acid, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of elastase by N-sulfonylaryl beta-lactams: anatomy of a stable acyl-enzyme complex.
Biochemistry, 37, 1998
|
|
1PYY
 
 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
1J7U
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Complex | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1WAV
 
 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
1J7L
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, MAGNESIUM ION | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
1SZC
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1GHF
 
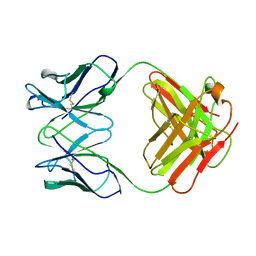 | | ANTI-ANTI-IDIOTYPE GH1002 FAB FRAGMENT | | Descriptor: | ANTI-ANTI-IDIOTYPE GH1002 FAB FRAGMENT | | Authors: | Ban, N, Day, J, Wang, X, Ferrone, S, McPherson, A. | | Deposit date: | 1995-11-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an anti-anti-idiotype shows it to be self-complementary.
J.Mol.Biol., 255, 1996
|
|
5QKO
 
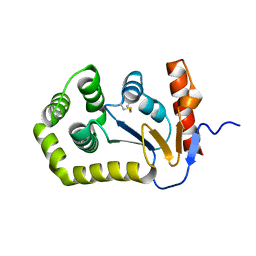 | |
5QL3
 
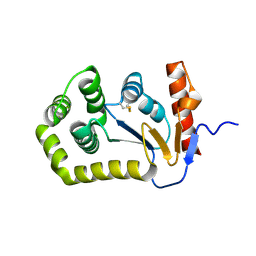 | |
5QLM
 
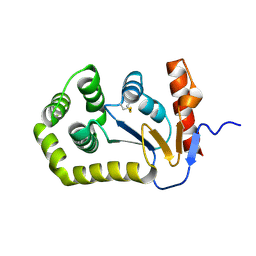 | |
5QM0
 
 | |
