8PN1
 
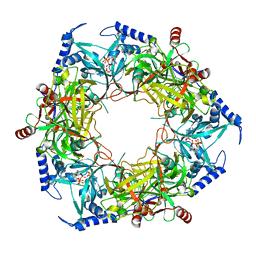 | | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
9F8G
 
 | |
3RHB
 
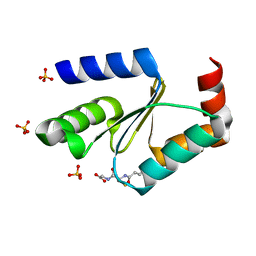 | | Crystal structure of the apo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, Glutaredoxin-C5, chloroplastic, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
3RHC
 
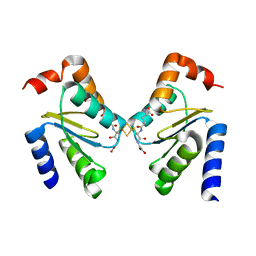 | | Crystal structure of the holo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Glutaredoxin-C5, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6NHT
 
 | |
7DIG
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7CZ0
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb92) | | Descriptor: | ACETATE ION, CACODYLATE ION, CACODYLIC ACID, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-09-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | An improved fluorescent protein tag and its nanobodies for membrane protein expression, stability assay, and purification
To Be Published
|
|
4EEU
 
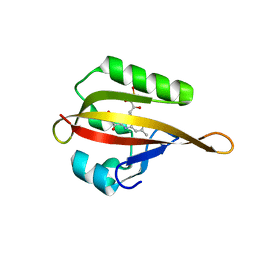 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EER
 
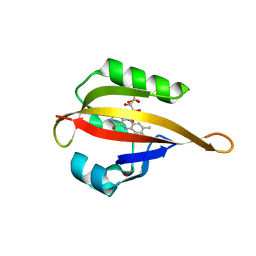 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EET
 
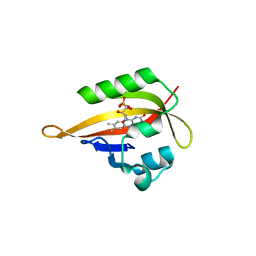 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EEP
 
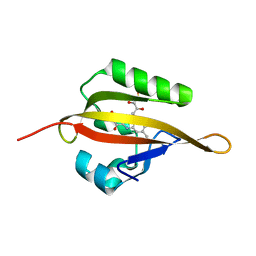 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4DE8
 
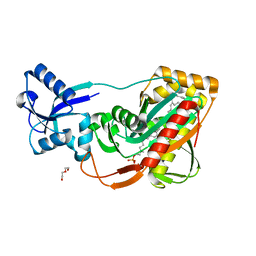 | | LytR-Cps2a-Psr family protein with bound octaprenyl monophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl dihydrogen phosphate, Cps2A, DI(HYDROXYETHYL)ETHER | | Authors: | Eberhardt, A, Hoyland, C.N, Vollmer, D.V, Bisle, S, Cleverley, R.M, Johnsborg, O, Havarstein, L.S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2012-01-20 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae.
Microb Drug Resist, 18, 2012
|
|
3C1Q
 
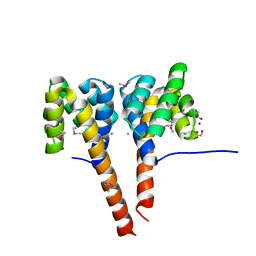 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Abendroth, J, Mitchell, D.D, Korotkov, K.V, Kreeger, A, Hol, W.G.J. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae
J.Struct.Biol., 166, 2009
|
|
2FL1
 
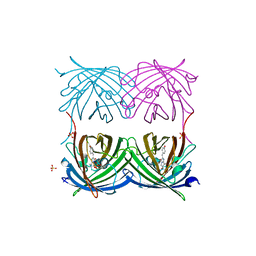 | | Crystal structure of red fluorescent protein from Zoanthus, zRFP574, at 2.4A resolution | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, V, Pletneva, N, Martynov, V, Tikhonova, T, Popov, B, Pletnev, S. | | Deposit date: | 2006-01-05 | | Release date: | 2007-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a red fluorescent protein from Zoanthus, zRFP574, reveals a novel chromophore
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4GPB
 
 | |
4DE9
 
 | | LytR-CPS2A-psr family protein YwtF (TagT) with bound octaprenyl pyrophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22E,26E)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, Putative transcriptional regulator ywtF | | Authors: | Eberhardt, A, Hoyland, C.N, Vollmer, D, Bisle, S, Cleverley, R.M, Johnsborg, O, Havarstein, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2012-01-20 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae.
Microb Drug Resist, 18, 2012
|
|
4EES
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
2HPW
 
 | | Green fluorescent protein from Clytia gregaria | | Descriptor: | Green fluorescent protein | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, S.E, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Green Fluorescent Protein from Clytia Gregaria at 1.55 A resolution
To be Published
|
|
3WUP
 
 | | Crystal Structure of the Ubiquitin-Binding Zinc Finger (UBZ) Domain of the Human DNA Polymerase Eta | | Descriptor: | CHLORIDE ION, DNA polymerase eta, GLYCEROL, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, S. | | Deposit date: | 2014-05-01 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 283, 2016
|
|
3ZUL
 
 | |
4BD8
 
 | | Bax domain swapped dimer induced by BimBH3 with CHAPS | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BAX, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
7UJ0
 
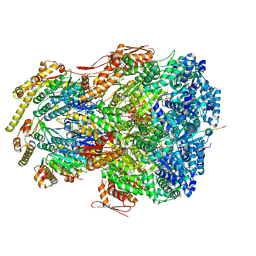 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
