4LEL
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCG-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.90000033 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LNT
 
 | | Crystal Structure of tRNA Proline (CGG) Bound to Codon CCC-U on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-07-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CGX
 
 | | Crystal structure of the N-terminal domain of the PA subunit of Thogoto virus polymerase (form 1) | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CHE
 
 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CGS
 
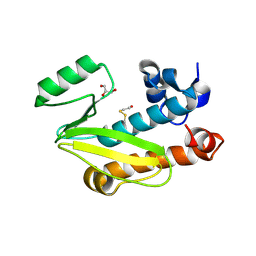 | | Crystal structure of the N-terminal domain of the PA subunit of Dhori virus polymerase | | Descriptor: | GLYCEROL, POLYMERASE SUBUNIT PA | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-26 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4IIL
 
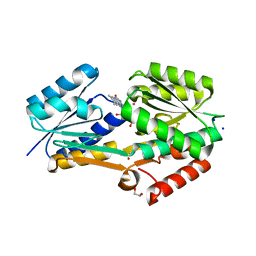 | | Crystal Structure of RfuA (TP0298) of T. pallidum Bound to Riboflavin | | Descriptor: | 1,2-ETHANEDIOL, Membrane lipoprotein TpN38(b), POTASSIUM ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes.
MBio, 4, 2013
|
|
8GCQ
 
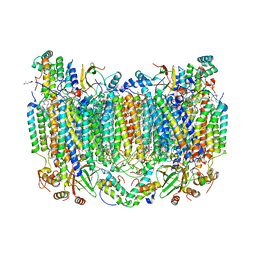 | | SFX structure of oxidized cytochrome c oxidase at 2.38 Angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ishigami, I, Yeh, S.-R, Rousseau, D.L. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural insights into functional properties of the oxidized form of cytochrome c oxidase.
Nat Commun, 14, 2023
|
|
6YUF
 
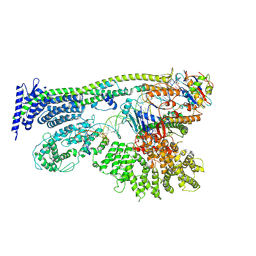 | | Cohesin complex with loader gripping DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit rad21, ... | | Authors: | Higashi, T.L, Eickhoff, P, Sousa, J.S, Costa, A, Uhlmann, F. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | A Structure-Based Mechanism for DNA Entry into the Cohesin Ring.
Mol.Cell, 79, 2020
|
|
7PQU
 
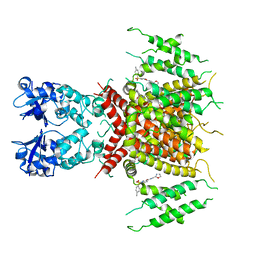 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
6FT3
 
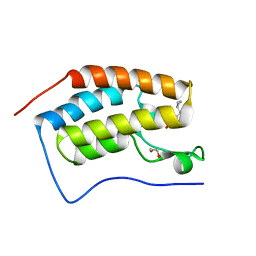 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FSY
 
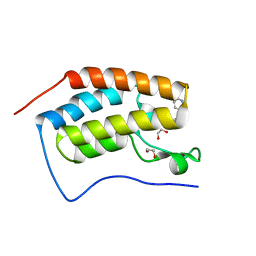 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
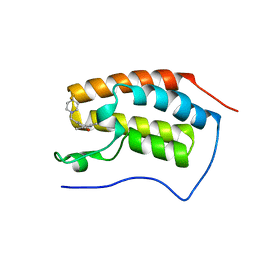 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6WBO
 
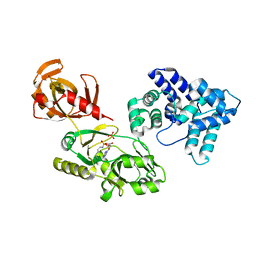 | |
4ZXG
 
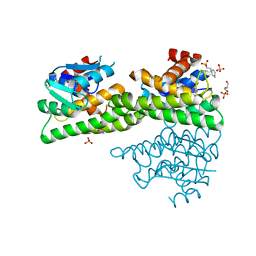 | | Ligandin binding site of PfGST | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Perbandt, M, Eberle, R, Betzel, C. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structures of Plasmodium falciparum GST complexes provide novel insights into the dimer-tetramer transition and a novel ligand-binding site.
J.Struct.Biol., 191, 2015
|
|
5X5S
 
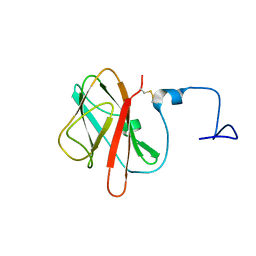 | |
1HJX
 
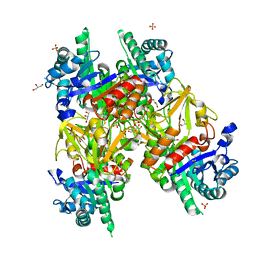 | | Ligand-induced signalling and conformational change of the 39 kD glycoprotein from human articular chondrocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3 LIKE PROTEIN 1, GLYCEROL, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
2Y2Z
 
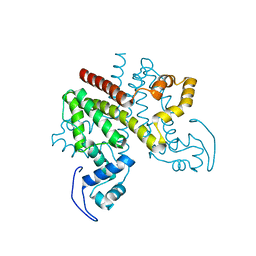 | | ligand-free form of TetR-like repressor SimR | | Descriptor: | PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
5CW0
 
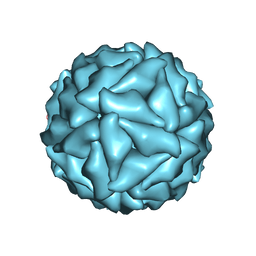 | |
8DPX
 
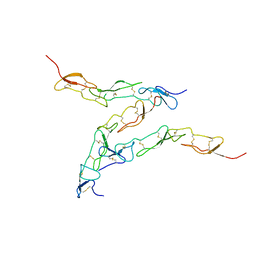 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
6V0B
 
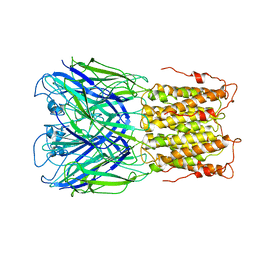 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1VRK
 
 | |
7L6Q
 
 | | Unliganded ELIC in styrene-maleic-acid nanodiscs at 2.5-Angstrom resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2020-12-23 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L6U
 
 | |
2LBD
 
 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO ALL-TRANS RETINOIC ACID | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Renaud, J.-P, Rochel, N, Ruff, M, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid.
Nature, 378, 1995
|
|
1KN4
 
 | | CATALYTIC ANTIBODY D2.3 COMPLEX | | Descriptor: | IG ANTIBODY D2.3 (HEAVY CHAIN), IG ANTIBODY D2.3 (LIGHT CHAIN), PARA-NITROPHENYL PHOSPHONOBUTANOYL D-ALANINE, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remarkable remote chiral recognition in a reaction mediated by a catalytic antibody.
J.Am.Chem.Soc., 124, 2002
|
|
