2OF0
 
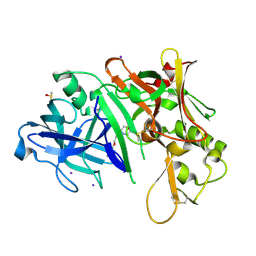 | | X-ray crystal structure of beta secretase complexed with compound 5 | | Descriptor: | (2S)-1-(2,5-dimethylphenoxy)-3-morpholin-4-ylpropan-2-ol, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-02 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to beta-Secretase.
J.Med.Chem., 50, 2007
|
|
2OHR
 
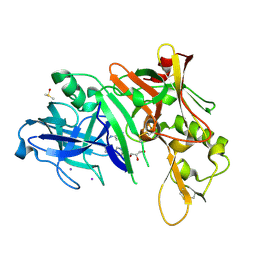 | | X-ray crystal structure of beta secretase complexed with compound 6a | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
2OHN
 
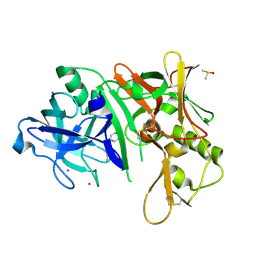 | |
1QBU
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R--(1ALPHA,5ALPHA,7BETA)]-3-[(CYCLOPROPHYLMETHYL)HEXAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPIN] METHYL-2-THIAZOLYLBENZAMIDE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
3ZM5
 
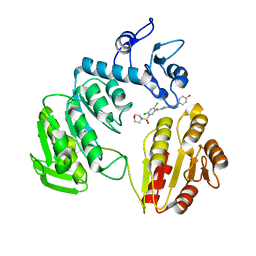 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
6T37
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T60
 
 | |
6T6N
 
 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
1X7A
 
 | | Porcine Factor IXa Complexed to 1-{3-[amino(imino)methyl]phenyl}-N-[4-(1H-benzimidazol-1-yl)-2-fluorophenyl]-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide | | Descriptor: | 1-{3-[AMINO(IMINO)METHYL]PHENYL}-N-[4-(1H-BENZIMIDAZOL-1-YL)-2-FLUOROPHENYL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOLE-5-CARBOXAMIDE, Coagulation Factor IX, light chain, ... | | Authors: | Alexander, R.S, Smallwood, A.M, Smallheer, J.M, Wang, J, Wang, S, Nakajima, S, Rossi, K.A, Barbera, F, Burdick, D, Luettgen, J.M. | | Deposit date: | 2004-08-13 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | SAR and factor IXa crystal structure of a dual inhibitor of factors IXa and Xa
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2OHS
 
 | | X-ray crystal structure of beta secretase complexed with compound 6b | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Patel, S. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase.
J.Med.Chem., 50, 2007
|
|
6RFW
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1039 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-[2-(2-fluorophenyl)ethyl]prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
6TMA
 
 | |
4ANM
 
 | | Complex of CK2 with a CDC7 inhibitor | | Descriptor: | 8-BROMANYL-2-[[(3S)-3-OXIDANYLPYRROLIDIN-1-YL]METHYL]-3H-[1]BENZOFURO[3,2-D]PYRIMIDIN-4-ONE, CASEIN KINASE II SUBUNIT ALPHA | | Authors: | Stout, T.J. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Xl413, a Potent and Selective Cdc7 Inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1XY9
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
4N07
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
2HB1
 
 | | Crystal Structure of PTP1B with Monocyclic Thiophene Inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K, Lee, J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5FX5
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1XY5
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
6RFN
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1018 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(3-methoxyphenyl)prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
1ZJ7
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
1ZLF
 
 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
4EDY
 
 | | Crystal structure of hH-PGDS with water displacing inhibitor | | Descriptor: | 4-[2-(hydroxymethyl)naphthalen-1-yl]-N-[2-(morpholin-4-yl)ethyl]benzamide, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Day, J.E, Thorarensen, A, Trujillo, J.I, Kiefer, J.R. | | Deposit date: | 2012-03-27 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Investigation of the binding pocket of human hematopoietic prostaglandin (PG) D2 synthase (hH-PGDS): a tale of two waters.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EKE
 
 | | Crystal structure of GPb in complex with DK11 | | Descriptor: | 3-(beta-D-glucopyranosyl)-6-pentylfuro[2,3-d]pyrimidin-2(3H)-one, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2012-04-09 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The binding of C5-alkynyl and alkylfurano[2,3-d]pyrimidine glucopyranonucleosides to glycogen phosphorylase b: Synthesis, biochemical and biological assessment.
Eur.J.Med.Chem., 54, 2012
|
|
4KFP
 
 | | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived Ureas as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[1-(tetrahydro-2H-pyran-4-yl)piperidin-4-yl]sulfonyl}benzyl)-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Bair, K.W, Baumeister, T, Ho, Y, Liederer, B.M, Liu, X, O'Brien, T, Oeh, J, Sampath, D, Skelton, N, Wang, L, Wang, W, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X. | | Deposit date: | 2013-04-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of 2,3-dihydro-1H-pyrrolo[3,4-c]pyridine-derived ureas as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5EKX
 
 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYLMETHIONINE AND FRAGMENT NB2E11 | | Descriptor: | 4-chloro-5-methylbenzene-1,2-diamine, NS5 METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
