6LTQ
 
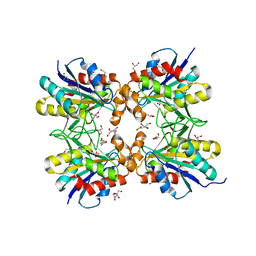 | | Crystal structure of pyrrolidone carboxyl peptidase from thermophilic keratin degrading bacterium Fervidobacterium islandicum AW-1 (FiPcp) | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Pyroglutamyl-peptidase I | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-01-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oxidized pyrrolidone carboxypeptidase from Fervidobacterium islandicum AW-1 reveals unique structural features for thermostability and keratinolysis.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
6TY7
 
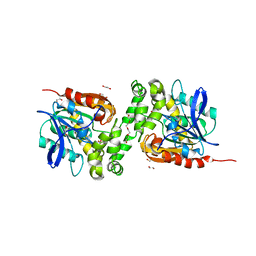 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
6TZI
 
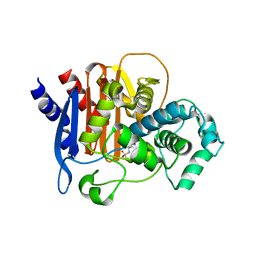 | | ADC-7 in complex with boronic acid transition state inhibitor PFC_001 | | Descriptor: | Beta-lactamase, phosphonooxy-[[4-[[2,2,2-tris(fluoranyl)ethylsulfonylamino]methyl]-1,2,3-triazol-1-yl]methyl]borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
5K0T
 
 | |
6TZF
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S17079 | | Descriptor: | 3-(1-{[hydroxy(phosphonooxy)boranyl]methyl}-1H-1,2,3-triazol-4-yl)benzoic acid, Beta-lactamase | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Banerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the murine LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state
Nature Communications, 2025
|
|
6E8Y
 
 | | Unliganded Human Glycerol 3-Phosphate Dehydrogenase | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Mydy, L.S, Gulick, A.M. | | Deposit date: | 2018-07-31 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human Glycerol 3-Phosphate Dehydrogenase: X-ray Crystal Structures That Guide the Interpretation of Mutagenesis Studies.
Biochemistry, 58, 2019
|
|
3K82
 
 | | Crystal Structure of the third PDZ domain of PSD-95 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disks large homolog 4, GLYCEROL, ... | | Authors: | Camara-Artigas, A, Gavira, J.A. | | Deposit date: | 2009-10-13 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4A X-ray structures
J.Struct.Biol., 170, 2010
|
|
5KM0
 
 | |
6GQO
 
 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 18) | | Descriptor: | 2-[4-(6,7-dimethoxyquinazolin-4-yl)oxy-2-methoxy-phenyl]-~{N}-(1-propan-2-ylpyrazol-4-yl)ethanamide, Vascular endothelial growth factor receptor 2 | | Authors: | Ogg, D.J, Schimpl, M, Hardy, C.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
1CED
 
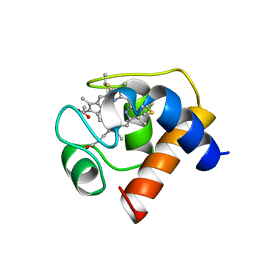 | | THE STRUCTURE OF CYTOCHROME C6 FROM MONORAPHIDIUM BRAUNII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Quacquarini, G, Walter, O, Diaz, A, Hervas, M, De La Rosa, M.A. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The solution structure of cytochrome c6 from the green alga Monoraphidium braunii
J.Biol.Inorg.Chem., 1, 1996
|
|
9FQS
 
 | | EGFR Exon20 insertion mutant NPG bound with Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-fluoranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-propanoylpyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-06-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery of STX-721, a Covalent, Potent, and Highly Mutant-Selective EGFR/HER2 Exon20 Insertion Inhibitor for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 68, 2025
|
|
6GKE
 
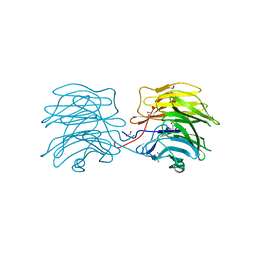 | | Aleuria aurantia lectin AAL N224Q mutant in complex with Fucalpha1-6GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Fucose-specific lectin, GLYCEROL, ... | | Authors: | Houser, J, Wimmerova, M, Herrera, H, Mehta, A.S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural dynamics and applications of a fucose-binding lectin with enhanced binding to alpha-1,6-linked core fucosylation
To Be Published
|
|
8BHE
 
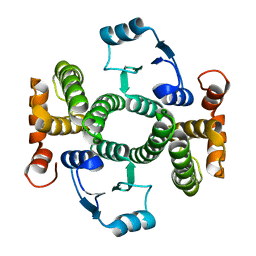 | | K141H and S142H double mutant of hGSTA1-1 | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2022-10-31 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Key Role in Catalysis and Enzyme Thermostability of a Conserved Helix H5 Motif of Human Glutathione Transferase A1-1.
Int J Mol Sci, 24, 2023
|
|
5HGK
 
 | |
1ZC5
 
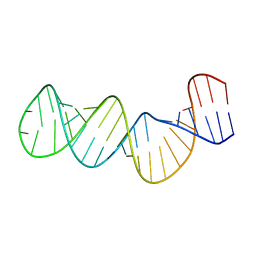 | | Structure of the RNA signal essential for translational frameshifting in HIV-1 | | Descriptor: | HIV-1 frameshift RNA signal | | Authors: | Gaudin, C, Mazauric, M.H, Traikia, M, Guittet, E, Yoshizawa, S, Fourmy, D. | | Deposit date: | 2005-04-11 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA Signal Essential for Translational Frameshifting in HIV-1
J.Mol.Biol., 349, 2005
|
|
5HIL
 
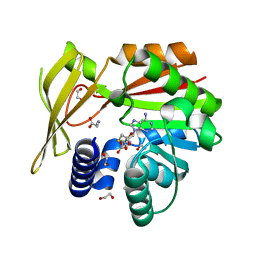 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
9JA8
 
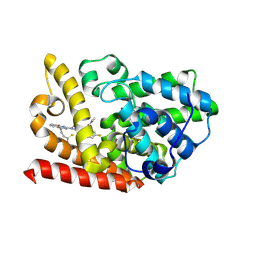 | | Crystal structure of human phosphodiesterase 10A in complex with N-(2-amino-2-thioxoethyl)-2-(3-(3-(dimethylcarbamoyl)-6-fluoroimidazo[1,2-a]pyridin-2-yl)azetidin-1-yl)quinoline-4-carboxamide | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Zhang, F.C, Huang, Y.Y, Luo, H.B, Guo, L. | | Deposit date: | 2024-08-24 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhalable Carbonyl Sulfide Donor-Hybridized Selective Phosphodiesterase 10A Inhibitor for Treating Idiopathic Pulmonary Fibrosis by Inhibiting Tumor Growth Factor-beta Signaling and Activating the cAMP/Protein Kinase A/cAMP Response Element-Binding Protein (CREB)/p53 Axis.
Acs Pharmacol Transl Sci, 8, 2025
|
|
7STB
 
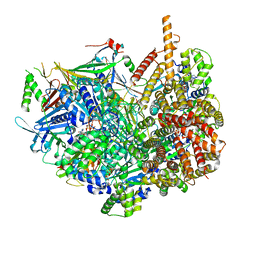 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5A6A
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with NGT | | Descriptor: | 1,2-ETHANEDIOL, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5J1X
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-5 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, Neuropilin-1, N~2~-(tert-butoxycarbonyl)-L-arginine | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
8XN2
 
 | | SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
7ST9
 
 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7MBY
 
 | | Human Cholecystokinin 1 receptor (CCK1R) Gq chimera (mGsqi) complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Mobbs, J.I, Belousoff, M.J, Danev, R, Thal, D.M, Sexton, P.M. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity.
Plos Biol., 19, 2021
|
|
5B07
 
 | | Lysozyme (denatured by DCl and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
