4B6M
 
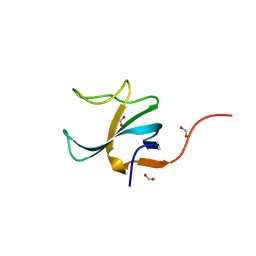 | |
2JWG
 
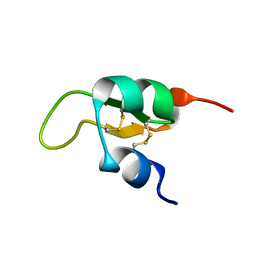 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
1R26
 
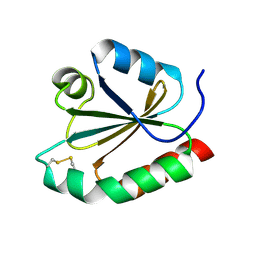 | | Crystal structure of thioredoxin from Trypanosoma brucei brucei | | Descriptor: | Thioredoxin | | Authors: | Friemann, R, Schmidt, H, Ramaswamy, S, Forstner, M, Krauth-Siegel, R.L, Eklund, H. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of thioredoxin from Trypanosoma brucei brucei
FEBS Lett., 554, 2003
|
|
1L8S
 
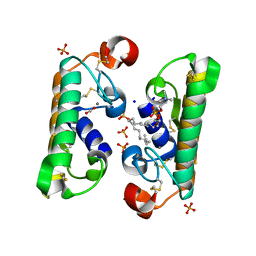 | |
3RQW
 
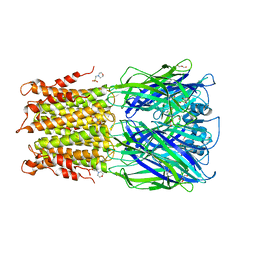 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3RQU
 
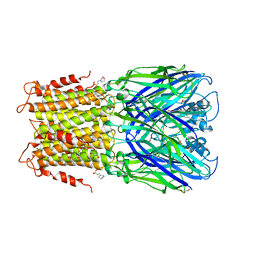 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3FVJ
 
 | |
1FX9
 
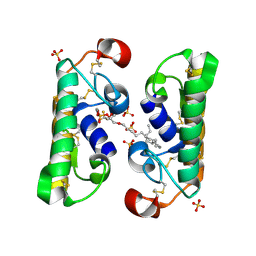 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + SULPHATE IONS) | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
1FXF
 
 | | CARBOXYLIC ESTER HYDROLASE COMPLEX (DIMERIC PLA2 + MJ33 INHIBITOR + PHOSPHATE IONS) | | Descriptor: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Pan, Y.H, Epstein, T.M, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Five coplanar anion binding sites on one face of phospholipase A2: relationship to interface binding.
Biochemistry, 40, 2001
|
|
5Z98
 
 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
7WH0
 
 | | structure of C elegans BCMO-1 | | Descriptor: | Beta-Carotene 15,15'-MonoOxygenase, FE (III) ION, GLYCEROL, ... | | Authors: | Pan, W, Liu, L. | | Deposit date: | 2021-12-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of Nonheme Iron Enzymes BCMO-1 and BCMO-2 from Caenorhabditis elegans .
Front Mol Biosci, 9, 2022
|
|
6SJQ
 
 | |
7WH1
 
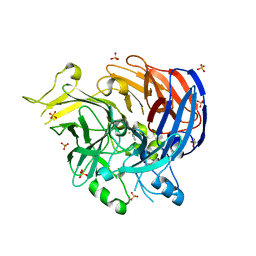 | | structure of C elegans BCMO-2 | | Descriptor: | Beta-Carotene 15,15'-MonoOxygenase, FE (III) ION, GLYCEROL, ... | | Authors: | Pan, W, Liu, L. | | Deposit date: | 2021-12-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Nonheme Iron Enzymes BCMO-1 and BCMO-2 from Caenorhabditis elegans .
Front Mol Biosci, 9, 2022
|
|
3OR7
 
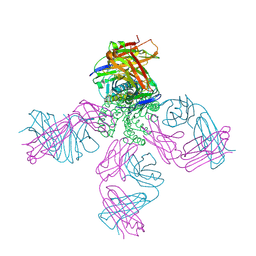 | | On the structural basis of modal gating behavior in K+channels - E71I | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Chakrapani, S, Cordero-Morales, J.F, Jogini, V, Pan, A.C, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2010-09-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis of modal gating behavior in K(+) channels.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6HEA
 
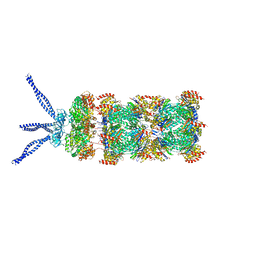 | | PAN-proteasome in state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.04 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE8
 
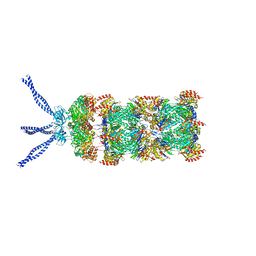 | | PAN-proteasome in state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.86 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HED
 
 | | PAN-proteasome in state 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HE9
 
 | | PAN-proteasome in state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HEC
 
 | | PAN-proteasome in state 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2J1Q
 
 | | Crystal structure of Trypanosoma cruzi arginine kinase | | Descriptor: | ARGININE KINASE, GLYCEROL | | Authors: | Fernandez, P, Haouz, A, Pereira, C.A, Aguilar, C, Alzari, P.M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Arginine Kinase.
Proteins, 69, 2007
|
|
6LK4
 
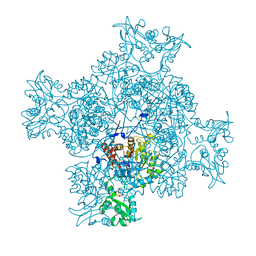 | | Crystal structure of GMP reductase from Trypanosoma brucei in complex with guanosine 5'-triphosphate | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Guanosine 5'-monophosphate Reductase, PHOSPHATE ION | | Authors: | Mase, H, Otani, T, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
4F57
 
 | |
3OR6
 
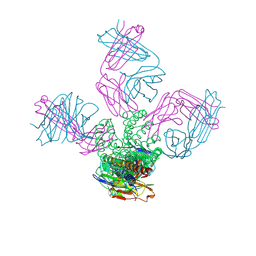 | | On the structural basis of modal gating behavior in K+channels - E71Q | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Chakrapani, S, Cordero-Morales, J.F, Jogini, V, Perozo, E. | | Deposit date: | 2010-09-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the structural basis of modal gating behavior in K(+) channels.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7AVM
 
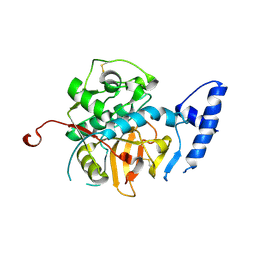 | | Crystal Structure of Pro-Rhodesain C150A | | Descriptor: | Cysteine protease, GLYCEROL | | Authors: | Johe, P, Jaenicke, E, Neuweiler, H, Schirmeister, T, Kersten, C, Hellmich, U. | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, interdomain dynamics, and pH-dependent autoactivation of pro-rhodesain, the main lysosomal cysteine protease from African trypanosomes.
J.Biol.Chem., 296, 2021
|
|
