4LE9
 
 | | Crystal structure of a chimeric c-Src-SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A, Martinez-Rodriguez, S, Ortiz-Salmeron, E, Martin-Garcia, J.M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops.
J.Struct.Biol., 186, 2014
|
|
7ZR2
 
 | | Crystal structure of a chimeric protein mimic of SARS-CoV-2 Spike HR1 in complex with HR2 | | Descriptor: | Spike protein S2', Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | | Authors: | Camara-Artigas, A, Gavira, J.A, Cano-Munoz, M, Polo-Megias, D, Conejero-Lara, F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel chimeric proteins mimicking SARS-CoV-2 spike epitopes with broad inhibitory activity.
Int.J.Biol.Macromol., 222, 2022
|
|
8AQO
 
 | | Streptavidin with a bisbiothinilated Fe4S4 cluster | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[[20-[2-[5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]ethanoylamino]-2$l^{3},4,12,14$l^{3},16,24,25$l^{3},27$l^{3}-octathia-1$l^{4},3$l^{4},13$l^{4},15$l^{4}-tetraferranonacyclo[11.11.1.1^{1,13}.1^{6,10}.1^{18,22}.0^{2,15}.0^{3,14}.0^{3,25}.0^{15,27}]octacosa-6(28),7,9,18,20,22(26)-hexaen-8-yl]amino]-2-oxidanylidene-ethyl]pentanamide, Streptavidin | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydrophobic probe bound to Streptavidin - 1
To Be Published
|
|
3PRG
 
 | | LIGAND BINDING DOMAIN OF HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Descriptor: | PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Uppenberg, J, Svensson, C, Jaki, M, Bertilsson, G, Jendeberg, L, Berkenstam, A. | | Deposit date: | 1998-08-24 | | Release date: | 1999-08-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand binding domain of the human nuclear receptor PPARgamma.
J.Biol.Chem., 273, 1998
|
|
6YBM
 
 | |
6VZO
 
 | |
6VZM
 
 | |
6VZN
 
 | |
6VZL
 
 | |
8AQY
 
 | | streptavidin mutant S112A with an iridium catalyst for CH activation | | Descriptor: | Streptavidin, tert-butyl 7'-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-1-chloranyl-2,3,4,5,6-pentamethyl-spiro[1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane-1,2'-3-aza-1-azonia-2$l^{8}-iridatricyclo[6.3.1.0^{4,12}]dodeca-1(11),4,6,8(12),9-pentaene]-3'-carboxylate | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | streptavidin mutant S112A with an iridium catalyst for CH activation
To Be Published
|
|
1M18
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | Histone H2A.1, Histone H2B.1, Histone H3.2, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
1M19
 
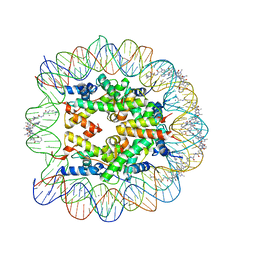 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
1M1A
 
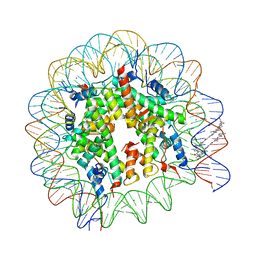 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
7AM3
 
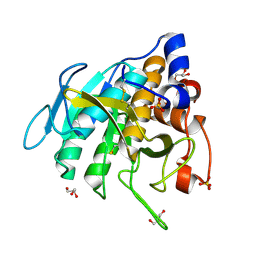 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM6
 
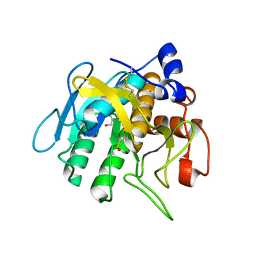 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM4
 
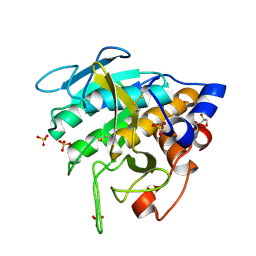 | |
7AM5
 
 | |
7AM7
 
 | |
6ZGJ
 
 | | GLIC pentameric ligand-gated ion channel, pH 5 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
4CGN
 
 | | Leishmania major N-myristoyltransferase in complex with a piperidinylindole inhibitor | | Descriptor: | 2-(4-fluorophenyl)-N-(3-piperidin-4-yl-1H-indol-5-yl)ethanamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
6ZGK
 
 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGD
 
 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
2C5C
 
 | |
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
