1LKB
 
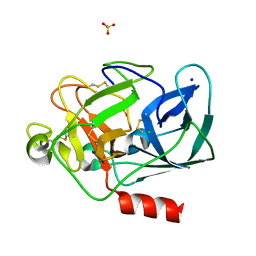 | | Porcine Pancreatic Elastase/Na-Complex | | Descriptor: | CHLORIDE ION, Elastase 1, SODIUM ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4FKD
 
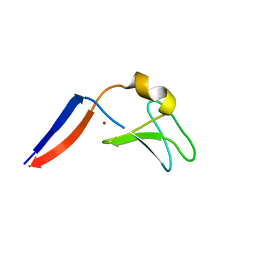 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
6BCA
 
 | |
3T95
 
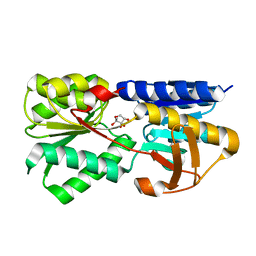 | | Crystal structure of LsrB from Yersinia pestis complexed with autoinducer-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein lsrB | | Authors: | Kavanaugh, J.S, Gakhar, L, Horswill, A.R. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of LsrB from Yersinia pestis complexed with autoinducer-2.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6EVG
 
 | |
1PVX
 
 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
5SYQ
 
 | | Solution structure of Aquifex aeolicus Aq1974 | | Descriptor: | Uncharacterized protein aq_1974 | | Authors: | Sachleben, J.R, Gawlak, G, Hoey, R.J, Liu, G, Joachimiak, A, Montelione, G.T, Koide, S, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aromatic claw: A new fold with high aromatic content that evades structural prediction.
Protein Sci., 26, 2017
|
|
6EGC
 
 | |
2C2T
 
 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-((7,8-dicarbaundecaboran-7-yl)methyl)-6-methylpyrimidine, a novel boron containing, nonclassical Antifolate | | Descriptor: | (S)-2,4-DIAMINO-5-((7,8-DICARBAUNDECABORAN-7-YL)METHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | leung, A.K.W, reynolds, R.C, riordan, J.M, borhani, D.W. | | Deposit date: | 2005-09-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
3I40
 
 | | Human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Chuprov-Netochin, R.N, Kuranova, I.P. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3UNX
 
 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6F0D
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group P1 with cadmium ions | | Descriptor: | CADMIUM ION, VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-19 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.90000749 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
1OW5
 
 | |
6FIE
 
 | | Crystallographic structure of calcium loaded Calbindin-D28K. | | Descriptor: | CALCIUM ION, Calbindin, THIOCYANATE ION | | Authors: | Noble, J.W, Almalki, R, Roe, S.M, Wagner, A, Dumanc, R, Atack, J.R. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The X-ray structure of human calbindin-D28K: an improved model.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1PH8
 
 | |
2C2S
 
 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-(1-o-carboranylmethyl)-6-methylpyrimidine, A novel boron containing, nonclassical Antifolate | | Descriptor: | 2,4-DIAMINO-5-(1-O-CARBORANYLMETHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Riordan, J.M, Borhani, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
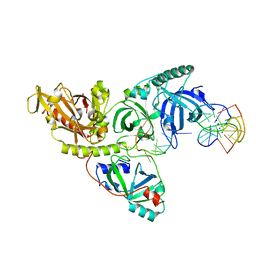
1PH9
 
 | |
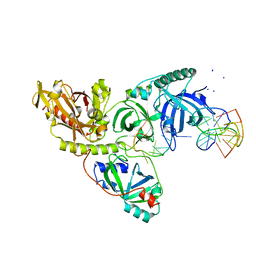
1PH6
 
 | |
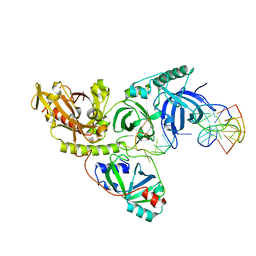
1PHJ
 
 | |
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
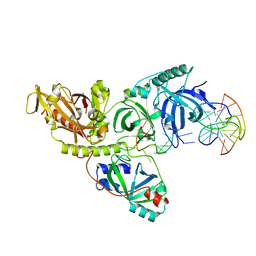
1PH1
 
 | |
1N2Y
 
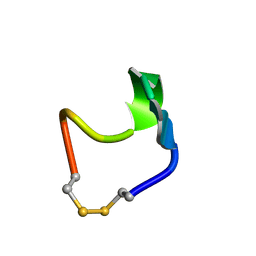 | | SOLUTION STRUCTURE OF SS-CYCLIZED CATESTATIN FRAGMENT FROM CHROMOGRANIN A | | Descriptor: | CATESTATIN | | Authors: | Preece, N.E, Nguyen, M, Mahata, M, Mahata, S.K, Mahapatra, N.R, Tsigelny, I, O'Connor, D.T. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine
release-inhibitory (catestatin) region of chromogranin A
Regul.Pept., 118, 2004
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
1J3G
 
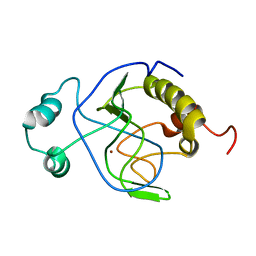 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
3GM7
 
 | |
