1H1O
 
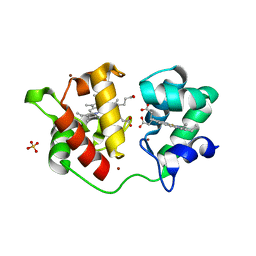 | | Acidithiobacillus ferrooxidans cytochrome c4 structure supports a complex-induced tuning of electron transfer | | Descriptor: | CYTOCHROME C-552, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abergel, C, Nitschke, W, Malarte, G, Bruschi, M, Claverie, J.-M, Guidici-Orticoni, M.-T. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Structure of Acidithiobacillus Ferrooxidans C(4)-Cytochrome. A Model for Complex-Induced Electron Transfer Tuning
Structure, 11, 2003
|
|
2VN9
 
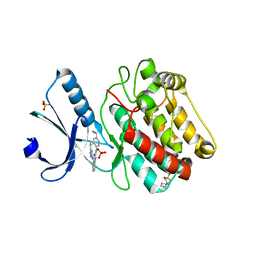 | | Crystal Structure of Human Calcium Calmodulin dependent Protein Kinase II delta isoform 1, CAMKD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II DELTA CHAIN, CHLORIDE ION, ... | | Authors: | Roos, A.K, Rellos, P, Salah, E, Pike, A.C.W, Fedorov, O, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
2Y4P
 
 | |
1VIH
 
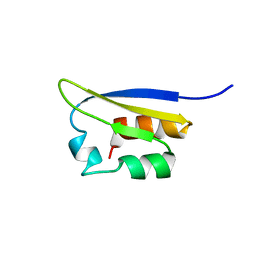 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VJ9
 
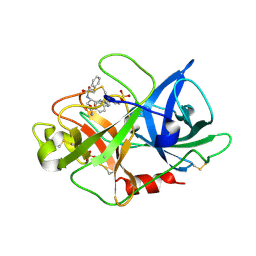 | | Urokinase Plasminogen Activator B-Chain-JT464 Complex | | Descriptor: | N-(BENZYLSULFONYL)-L-SERYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-O-BENZYL-L-SERINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
2BRH
 
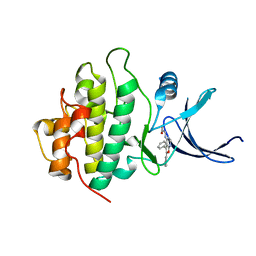 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | N-(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)GLYCINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2C3K
 
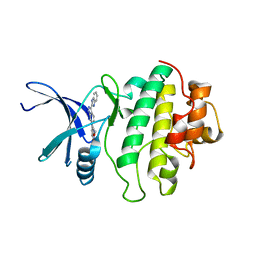 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 4-[3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOL-6-YL]-2-METHOXYPHENOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
1H92
 
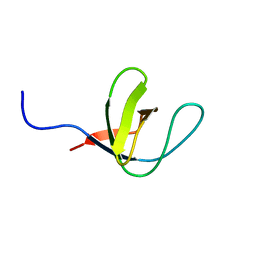 | | SH3 domain of human Lck tyrosine kinase | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Schweimer, K, Hoffmann, S, Friedrich, U, Biesinger, B, Roesch, P, Sticht, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-10-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck
Biochemistry, 41, 2002
|
|
1HHL
 
 | |
1W50
 
 | | Apo Structure of BACE (Beta Secretase) | | Descriptor: | BETA-SECRETASE 1, IODIDE ION | | Authors: | Patel, S, Vuillard, L, Cleasby, A, Murray, C.W, Yon, J. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Apo and Inhibitor Complex Structures of Bace (Beta-Secretase)
J.Mol.Biol., 343, 2004
|
|
2BHE
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 5-BROMO-INDIRUBINE | | Descriptor: | (2Z)-5'-BROMO-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE AMMONIATE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-10 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
2BL5
 
 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | Descriptor: | MGC83862 PROTEIN | | Authors: | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | Deposit date: | 2005-03-01 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
1W8J
 
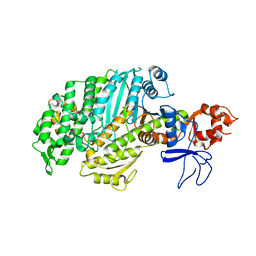 | |
2BZ2
 
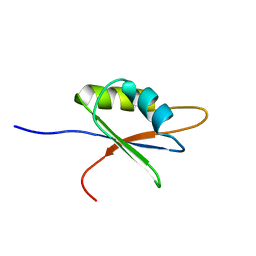 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
2BP2
 
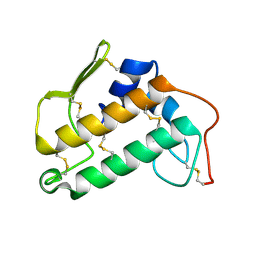 | | THE STRUCTURE OF BOVINE PANCREATIC PROPHOSPHOLIPASE A2 AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Vannes, G.J.H, Kalk, K.H, Brandenburg, N.P, Hol, W.G.J, Drenth, J. | | Deposit date: | 1981-06-05 | | Release date: | 1981-07-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Bovine Pancreatic Prophospholipase A2 at 3.0 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1W08
 
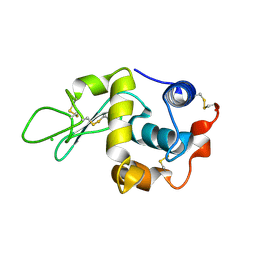 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
2C6U
 
 | |
3SQQ
 
 | | CDK2 in complex with inhibitor RC-3-96 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-methylbenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-07-06 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
1W4O
 
 | | Binding of Nonnatural 3'-Nucleotides to Ribonuclease A | | Descriptor: | PANCREATIC RIBONUCLEASE A, URACIL ARABINOSE-3'-PHOSPHATE | | Authors: | Jenkins, C.L, Thiyagarajan, N, Sweeney, R.Y, Guy, M.P, Kelemen, B.R, Acharya, K.R, Raines, R.T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of Non-Natural 3'-Nucleotides to Ribonuclease A
FEBS J., 272, 2005
|
|
1VZO
 
 | | The structure of the N-terminal kinase domain of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein | | Descriptor: | BETA-MERCAPTOETHANOL, RIBOSOMAL PROTEIN S6 KINASE ALPHA 5, SULFATE ION | | Authors: | Smith, K.J, Carter, P.S, Bridges, A, Horrocks, P, Lewis, C, Pettman, G, Clarke, A, Brown, M, Hughes, J, Wilkinson, M, Bax, B, Reith, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Msk1 Reveals a Novel Autoinhibitory Conformation for a Dual Kinase Protein
Structure, 12, 2004
|
|
2CJW
 
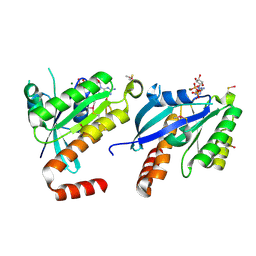 | | Crystal structure of the small GTPase Gem (GemDNDCaM) in complex to Mg.GDP | | Descriptor: | GTP-BINDING PROTEIN GEM, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Splingard, A, Menetrey, J, Perderiset, M, Cicolari, J, Hamoudi, F, Cabanie, L, El Marjou, A, Wells, A, Houdusse, A, de Gunzburg, J. | | Deposit date: | 2006-04-09 | | Release date: | 2006-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of the Gem Gtpase.
J.Biol.Chem., 282, 2007
|
|
2C9V
 
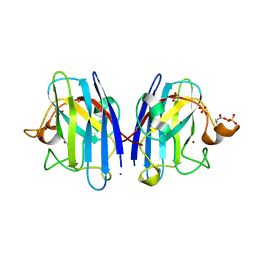 | | Atomic resolution structure of Cu-Zn Human Superoxide dismutase | | Descriptor: | COPPER (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2CEN
 
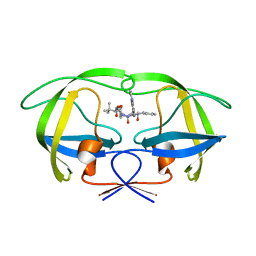 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
1W0G
 
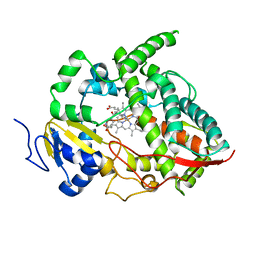 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
2CMM
 
 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
