8E2C
 
 | |
1Y7Y
 
 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
1I8O
 
 | |
1QLU
 
 | |
3ETY
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETW
 
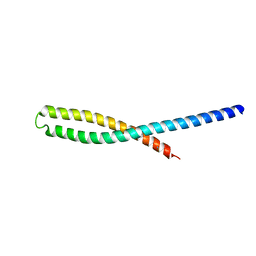 | | Crystal Structure of bacterial adhesin FadA | | Descriptor: | Adhesin A, THIOCYANATE ION | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
1LW2
 
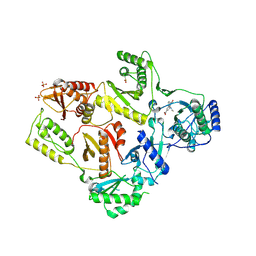 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
5NR9
 
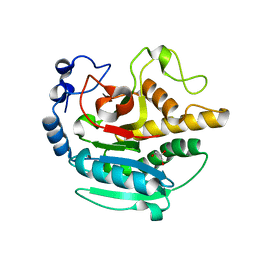 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | Descriptor: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6YA0
 
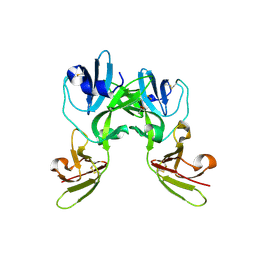 | | Crystal structure of TSWV glycoprotein N ectodomain (Trypsin treated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, AMMONIUM ION, ... | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8EGU
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
1E0O
 
 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
4ADS
 
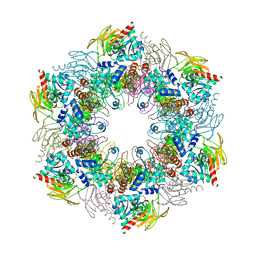 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
8ES0
 
 | |
8ERZ
 
 | |
8ERY
 
 | |
8ES2
 
 | |
8ES3
 
 | |
8ES1
 
 | |
3ETX
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETZ
 
 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
5NBK
 
 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6YK8
 
 | | OTU-like deubiquitinase from Legionella -Lpg2529 | | Descriptor: | PLATINUM (II) ION, Uncharacterized protein | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection.
Elife, 9, 2020
|
|
5O40
 
 | | SOD1 bound to Ebselen | | Descriptor: | N-phenyl-2-selanylbenzamide, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Capper, M.J, Wright, G.S.A, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The cysteine-reactive small molecule ebselen facilitates effective SOD1 maturation.
Nat Commun, 9, 2018
|
|
