7O0G
 
 | |
6NZ2
 
 | |
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
1HD7
 
 | | A Second Divalent Metal Ion in the Active Site of a New Crystal Form of Human Apurinic/Apyridinimic Endonuclease, Ape1, and its Implications for the Catalytic Mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-11-09 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
8J9A
 
 | |
2JZ3
 
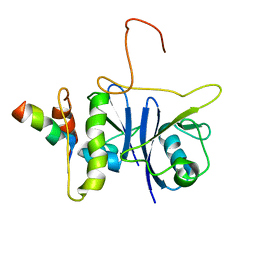 | | SOCS box elonginBC ternary complex | | Descriptor: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | Deposit date: | 2007-12-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
1K5M
 
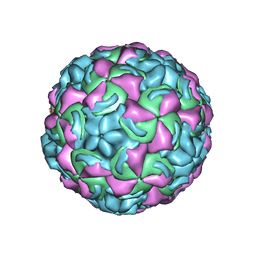 | | Crystal Structure of a Human Rhinovirus Type 14:Human Immunodeficiency Virus Type 1 V3 Loop Chimeric Virus MN-III-2 | | Descriptor: | CHIMERA OF HRV14 COAT PROTEIN VP2 (P1B) AND the V3 loop of HIV-1 gp120, COAT PROTEIN VP1 (P1D), COAT PROTEIN VP3 (P1C), ... | | Authors: | Ding, J, Smith, A.D, Geisler, S.C, Ma, X, Arnold, G.F, Arnold, E. | | Deposit date: | 2001-10-11 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Human Rhinovirus
that Displays Part of the HIV-1 V3 Loop and
Induces Neutralizing Antibodies against
HIV-1
Structure, 10, 2002
|
|
5O1T
 
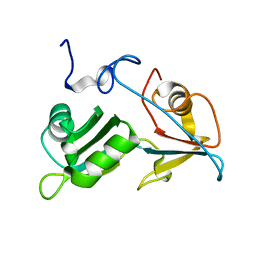 | |
7SSG
 
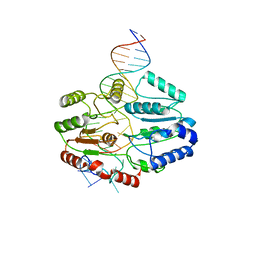 | | Mfd DNA complex | | Descriptor: | DNA (5'-D(P*TP*GP*GP*CP*GP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*GP*CP*CP*TP*CP*GP*CP*TP*GP*CP*CP*A)-3'), Transcription-repair-coupling factor | | Authors: | Oakley, A.J, Xu, Z.-Q. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Mechanism of transcription modulation by the transcription-repair coupling factor.
Nucleic Acids Res., 50, 2022
|
|
8VAR
 
 | |
8VAL
 
 | |
8VAQ
 
 | |
8VAP
 
 | |
8VAM
 
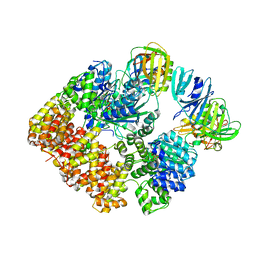 | |
8VAS
 
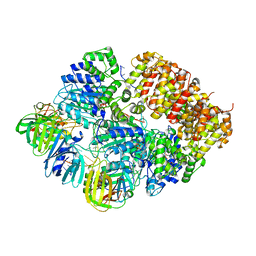 | |
8VAN
 
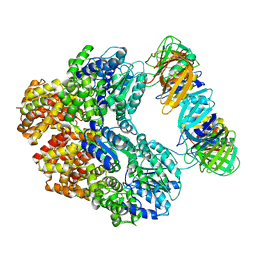 | | Structure of the E. coli clamp loader bound to the beta clamp in an Initial-Binding conformation | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Landeck, J.T, Pajak, J, Kelch, B.A. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
J.Biol.Chem., 300, 2024
|
|
7KKU
 
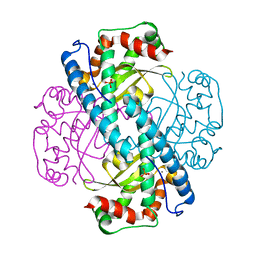 | | X-ray Counterpart to Neutron Structure of Oxidized Human MnSOD | | Descriptor: | MANGANESE (III) ION, PHOSPHATE ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KKS
 
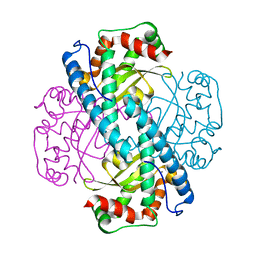 | | Neutron structure of Oxidized Human MnSOD | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
1HNI
 
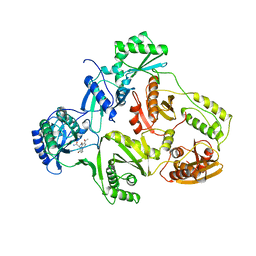 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
8CR8
 
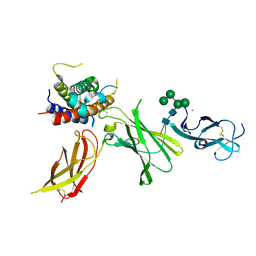 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6NUC
 
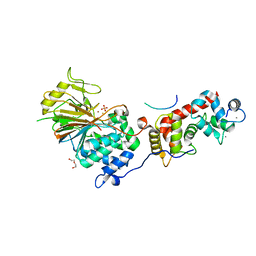 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
3PQY
 
 | | Crystal Structure of 6218 TCR in complex with the H2Db-PA224 | | Descriptor: | 10-mer peptide from RNA-directed RNA polymerase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-11-28 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6NUU
 
 | | Structure of Calcineurin mutant in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-02-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
6NUF
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
4QJD
 
 | |
