8OOO
 
 | |
8OOQ
 
 | |
8JHP
 
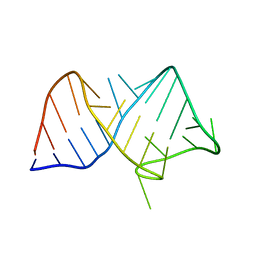 | |
7Q03
 
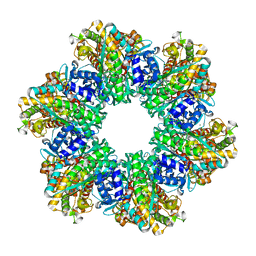
 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the close state with NADP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, GLYCEROL, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
7Q07
 
 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the open state with NADP and tartrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ketol-Acid Reductoisomerase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
7PX1
 
 | | Conotoxin from Conus mucronatus | | Descriptor: | CADMIUM ION, Conus mucronatus, IODIDE ION | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3.
Plos Biol., 21, 2023
|
|
5N1Q
 
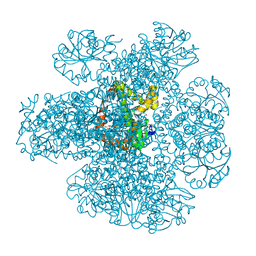
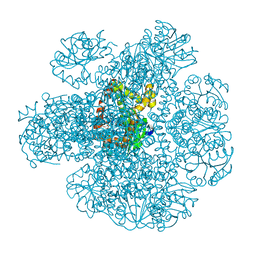
 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTHERMOCOCCUS THERMOLITHOTROPHICUS AT 1.9 A RESOLUTION | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
4YXB
 
 | | FliM(SPOA)::FliN fusion protein | | Descriptor: | Ambiguous peptide density, Flagellar motor switch protein FliM,Flagellar motor switch protein FliN, FliM::FliN fragment, ... | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
5CR1
 
 | | Crystal structure of TTR/resveratrol/T4 complex | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, RESVERATROL, Transthyretin | | Authors: | Zanotti, G, Florio, P, Folli, C, Cianci, M, Del Rio, D, Berni, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-amyloidogenic Activity of Natural Polyphenols and Their Metabolites.
J.Biol.Chem., 290, 2015
|
|
8TW6
 
 | | TCR in nanodisc ND-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, T cell receptor beta variable 6-5,T cell receptor beta chain MC.7.G5,MCHERRY, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
8TW4
 
 | | TCR in nanodisc ND-I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
3S9E
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
3RDP
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with N-METHYL-FHBT | | Descriptor: | 6-[(2R)-2-(fluoromethyl)-3-hydroxy-propyl]-1,5-dimethyl-pyrimidine-2,4-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
1H8Z
 
 | | Crystal structure of the class D beta-lactamase OXA-13 | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | Deposit date: | 2001-02-17 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
5ZNU
 
 | | Structure of omega conotoxin Bu8 | | Descriptor: | Omega-conotoxin-like Bu8 | | Authors: | Liu, X, Jiang, L. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega conotoxin bu8
To Be Published
|
|
9C3E
 
 | | TCR - CD3 complex bound to HLA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Cancer/testis antigen 1,Beta-2-microglobulin,MHC class I antigen, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-05-31 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
9BBC
 
 | | TCR GDN detergent micelle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-04-05 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
3H14
 
 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3HMU
 
 | | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, class III, CHLORIDE ION, ... | | Authors: | Toro, R, Bonanno, J.B, Ramagopal, U, Freeman, J, Bain, K.T, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi
To be Published
|
|
4WO0
 
 | | Crystal structure of transthyretin in complex with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Folli, C, Berni, R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1CAR
 
 | | I-CARRAGEENAN. MOLECULAR STRUCTURE AND PACKING OF POLYSACCHARIDE DOUBLE HELICES IN ORIENTED FIBRES OF DIVALENT CATION SALTS | | Descriptor: | 4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Iota-carrageenan: molecular structure and packing of polysaccharide double helices in oriented fibres of divalent cation salts.
J.Mol.Biol., 90, 1974
|
|
4WNJ
 
 | | Crystal structure of Transthyretin-quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Berni, R, Folli, C. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3F0T
 
 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with N-methyl-DHBT | | Descriptor: | N-Methyl-6-(1,3-dihydroxy-isobutyl)thymine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2008-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
1M22
 
 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
