1L5X
 
 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, Survival protein E | | Authors: | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | Deposit date: | 2002-03-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
1PSD
 
 | |
1CS1
 
 | |
3KMZ
 
 | | Crystal structure of RARalpha ligand binding domain in complex with the inverse agonist BMS493 and a corepressor fragment | | Descriptor: | 4-{(E)-2-[5,5-dimethyl-8-(phenylethynyl)-5,6-dihydronaphthalen-2-yl]ethenyl}benzoic acid, GLYCEROL, Nuclear receptor corepressor 1, ... | | Authors: | Bourguet, W, le Maire, A. | | Deposit date: | 2009-11-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1R43
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri (selenomethionine substituted protein) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-AMINO ISOBUTYRATE, ... | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-03 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278
|
|
1R3N
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-AMINO ISOBUTYRATE, ZINC ION, beta-alanine synthase | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278, 2003
|
|
2XC0
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4--DICARBOXYLIC ACID 3-[(3-FLUORO-4-METHOXY-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
4GAD
 
 | |
2XBY
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHYLCARBAMOYLMETHYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Thomi, S, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XBX
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XBW
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-SULFAMOYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Thomi, S, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
2XC5
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(4-CHLORO-3-FLUORO-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
1EC2
 
 | | HIV-1 protease in complex with the inhibitor BEA428 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-PYRIDIN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
7ZR0
 
 | | CryoEM structure of HSP90-CDC37-BRAF(V600E) complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Oberoi, J, Pearl, L.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation.
Nat Commun, 13, 2022
|
|
2GET
 
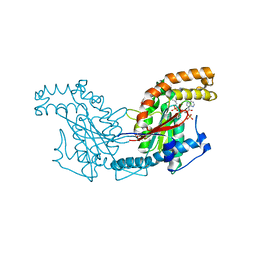 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-I (LT) | | Descriptor: | GLYCEROL, Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1AQZ
 
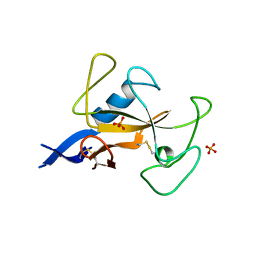 | |
2GEU
 
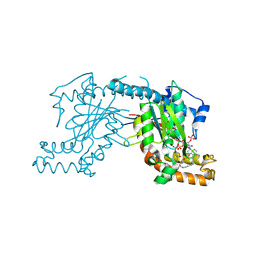 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-II (RT) | | Descriptor: | Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GEV
 
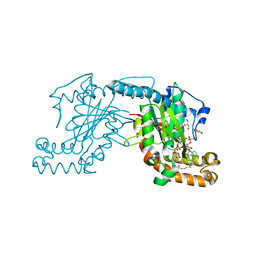 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-II (LT) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Pantothenate kinase, ... | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2JQV
 
 | |
2XBV
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-(2,2-DIFLUORO-ETHYL)-PYRROLIDINE-3,4-DICARBOXYLIC ACID 3-[(5-CHLORO-PYRIDIN-2-YL)-AMIDE]-4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
1PYG
 
 | |
1POS
 
 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1XKP
 
 | | Crystal structure of the virulence factor YopN in complex with its heterodimeric chaperone SycN-YscB | | Descriptor: | Chaperone protein sycN, Chaperone protein yscB, putative membrane-bound Yop targeting protein YopN | | Authors: | Schubot, F.D, Jackson, M.W, Penrose, K.J, Cherry, S, Tropea, J.E, Plano, G.V, Waugh, D.S. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis.
J.Mol.Biol., 346, 2005
|
|
1XL3
 
 | | Complex structure of Y.pestis virulence Factors YopN and TyeA | | Descriptor: | Secretion control protein, protein type A | | Authors: | Schubot, F.D, Jackson, M.W, Penrose, K.J, Cherry, S, Tropea, J.E, Plano, G.V, Waugh, D.S. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis.
J.Mol.Biol., 346, 2005
|
|
2GEZ
 
 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
