7AID
 
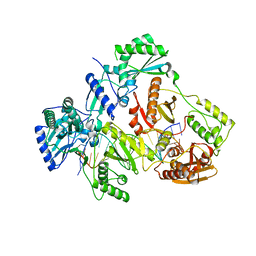 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
8SP3
 
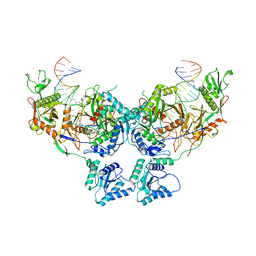 | | Asymmetric dimer of MapSPARTA bound with gRNA/tDNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-01 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
6VR4
 
 | |
8SP0
 
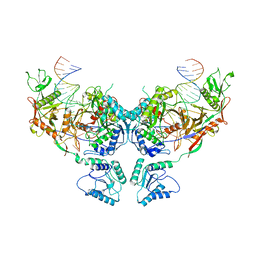 | | Symmetric dimer of MapSPARTA bound with gRNA/tDNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-01 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
4JRP
 
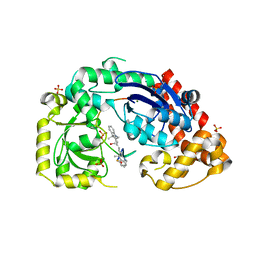 | | Structure of E. coli Exonuclease I in complex with a 5cy-dT13 oligonucleotide | | Descriptor: | 5cy-dT13, Exodeoxyribonuclease I, GLYCEROL, ... | | Authors: | Bell, C.E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Escherichia coli exonuclease I in complex with single-stranded DNA provide insights into the mechanism of processive digestion.
Nucleic Acids Res., 41, 2013
|
|
5UVQ
 
 | |
2XY8
 
 | |
4JRQ
 
 | |
8YCM
 
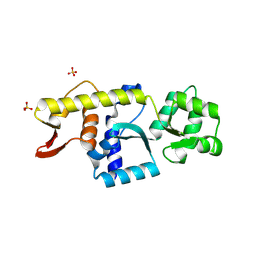 | | Monomeric Human STK19 | | Descriptor: | Isoform 4 of Inactive serine/threonine-protein kinase 19, SULFATE ION | | Authors: | Li, J, Ma, X, Dong, Z. | | Deposit date: | 2024-02-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mutations found in cancer patients compromise DNA binding of the winged helix protein STK19.
Sci Rep, 14, 2024
|
|
6G1N
 
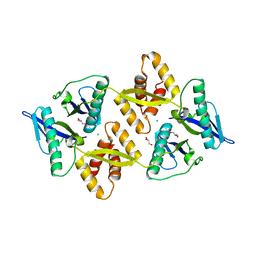 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
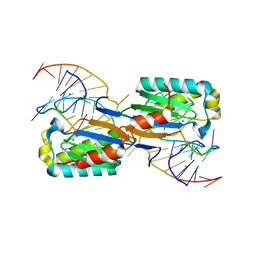
6FI8
 
 | |
6G3X
 
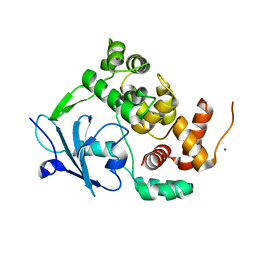 | |
6G1C
 
 | |
5ZCW
 
 | | Structure of the Methanosarcina mazei class II CPD-photolyase in complex with intact, phosphodiester linked, CPD-lesion | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*(TTD)P*CP*GP*CP*GP*CP*AP*A)-3', 5'-D(*TP*GP*CP*GP*CP*GP*AP*AP*GP*CP*CP*GP*AP*T)-3', ACETATE ION, ... | | Authors: | Maestre-Reyna, M, Bessho, Y. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
IUCrJ, 5, 2018
|
|
6G3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
8FTH
 
 | | Chaetomium thermophilum SETX - NPPC internal deletion | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
8FTM
 
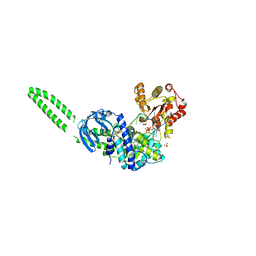 | | Setx-ssRNA-ADP-SO4 complex | | Descriptor: | 5'-3' RNA helicase-like protein, ADENOSINE-5'-DIPHOSPHATE, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Williams, R.S, Appel, C.D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Sen1 architecture: RNA-DNA hybrid resolution, autoregulation, and insights into SETX inactivation in AOA2.
Mol.Cell, 83, 2023
|
|
6G26
 
 | | The crystal structure of the Burkholderia pseudomallei HicAB complex | | Descriptor: | 1,2-ETHANEDIOL, HicA, HicB, ... | | Authors: | Winter, A.J, Isupov, M.N, Williams, C, Crump, M.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J. Biol. Chem., 293, 2018
|
|
5EYQ
 
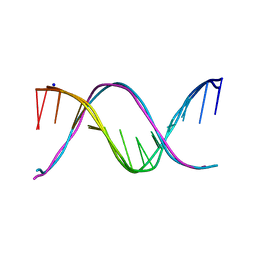 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pnna) | | Descriptor: | Complementary strand, Pribnow box template strand, SODIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-25 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
5Z00
 
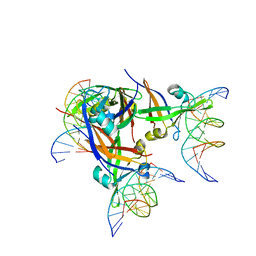 | | AtVAL1 B3 domain in complex with 15bp-DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*T)-3'), ... | | Authors: | Wu, B.X, Zhang, M.M. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
3DJL
 
 | |
6QVF
 
 | |
5TLB
 
 | | Scabin toxin from Streptomyces scabies in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Scabin | | Authors: | Lyons, B, Dutta, D, Ravulapalli, R, Merrill, A.R. | | Deposit date: | 2016-10-10 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of the catalytic signature of Scabin toxin, a DNA-targeting ADP-ribosyltransferase.
Biochem. J., 475, 2018
|
|
6QVI
 
 | |
