1IX5
 
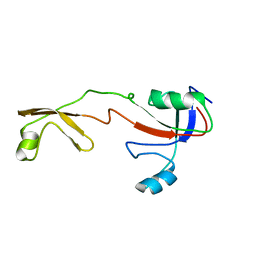 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
6B17
 
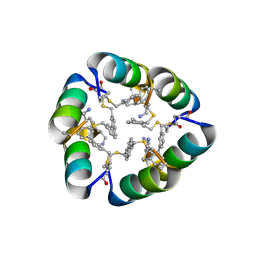 | | Design of a short thermally stable alpha-helix embedded in a macrocycle | | Descriptor: | 3,3'-dimethyl-1,1'-biphenyl, Capped-strapped peptide | | Authors: | Wu, H, Acharyya, A, Wu, Y, Liu, L, Jo, H, Gai, F, DeGrado, W.F. | | Deposit date: | 2017-09-17 | | Release date: | 2018-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Chembiochem, 19, 2018
|
|
1DZ1
 
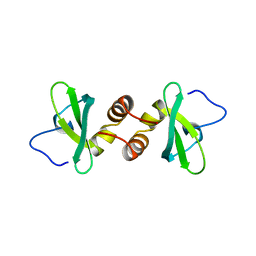 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | Descriptor: | MODIFIER 1 PROTEIN | | Authors: | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | Deposit date: | 2000-02-11 | | Release date: | 2000-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
1E9J
 
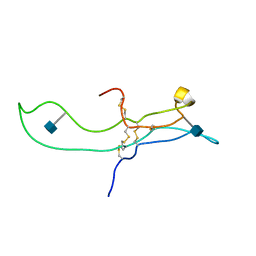 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [INCLUDING A SINGLE GLCNAC RESIDUE AT ASN52 AND ASN78] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-10-18 | | Release date: | 2002-07-25 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-linked glycans on the 3D structure of the free alpha-subunit of human chorionic gonadotropin.
Biochemistry, 39, 2000
|
|
1TTL
 
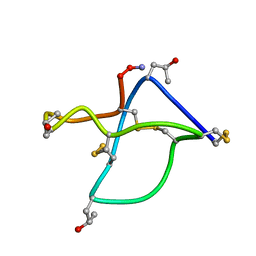 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
2MMC
 
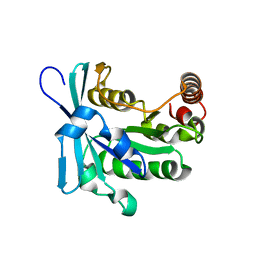 | | Nucleotide-free human ran gtpase | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of Ran GTPase Determines C-terminal Tail Conformational Dynamics.
To be Published
|
|
2MRK
 
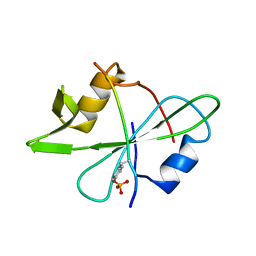 | | Fyn SH2 domain in complex with the natural inhibitory phosphotyrosine peptide | | Descriptor: | C-terminal Tyrosine-protein kinase Fyn, Tyrosine-protein kinase Fyn | | Authors: | Huculeci, R, Buts, L, Lenaerts, A.J, van Nuland, N. | | Deposit date: | 2014-07-09 | | Release date: | 2015-07-15 | | Last modified: | 2016-12-14 | | Method: | SOLUTION NMR | | Cite: | Dynamically Coupled Residues within the SH2 Domain of FYN Are Key to Unlocking Its Activity.
Structure, 24, 2016
|
|
2MRJ
 
 | |
1Q60
 
 | | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hayashi, F, Hirota, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA
To be published
|
|
2NB2
 
 | | Nigellin-1.1 | | Descriptor: | nigellin-1.1 | | Authors: | Bozin, T.N, Bocharov, E.V, Rogozhin, E.A. | | Deposit date: | 2016-01-20 | | Release date: | 2017-01-25 | | Method: | SOLUTION NMR | | Cite: | A novel type of hairpin-like defense peptides from blackseed (Nigella sativa L.) contains three disulfide bridges
To be Published
|
|
1MTX
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF MARGATOXIN BY 1H, 13C, 15N TRIPLE-RESONANCE NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MARGATOXIN | | Authors: | Johnson, B.A, Stevens, S.P, Williamson, J.M. | | Deposit date: | 1994-12-27 | | Release date: | 1995-11-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of margatoxin by 1H, 13C, 15N triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1OIG
 
 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
1OVY
 
 | |
1ONB
 
 | |
1OQJ
 
 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
1OK0
 
 | | Crystal Structure of Tendamistat | | Descriptor: | ALPHA-AMYLASE INHIBITOR HOE-467A, CHLORIDE ION, GLYCEROL | | Authors: | Koenig, V, Vertesy, L, Schneider, T.R. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Crystal Structure of the Alpha-Amylase Inhibitor Tendamistat at 0.93 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3ZOB
 
 | | Solution structure of chicken Engrailed 2 homeodomain | | Descriptor: | HOMEOBOX PROTEIN ENGRAILED-2 | | Authors: | Carlier, L, Balayssac, S, Cantrelle, F.X, Khemtemourian, L, Chassaing, G, Joliot, A, Lequin, O. | | Deposit date: | 2013-02-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Investigation of Homeodomain Membrane Translocation Properties: Insights from the Structure Determination of Engrailed-2 Homeodomain in Aqueous and Membrane-Mimetic Environments.
Biophys.J., 105, 2013
|
|
4AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
4AEN
 
 | | HLA-DR1 with covalently linked CLIP106-120 in reversed orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
4A4U
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*GP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4S
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*CP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4AH2
 
 | | HLA-DR1 with covalently linked CLIP106-120 in canonical orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN,DRB1-1 BETA CHAIN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
1QD7
 
 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1QMR
 
 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | Descriptor: | MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
4BD4
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, mutant H43F | | Descriptor: | GLYCEROL, SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Awad, W, Saraboji, K, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
