1MMO
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL NON-HAEM IRON HYDROXYLASE THAT CATALYSES THE BIOLOGICAL OXIDATION OF METHANE | | Descriptor: | ACETIC ACID, FE (III) ION, METHANE MONOOXYGENASE HYDROLASE (ALPHA CHAIN), ... | | Authors: | Rosenzweig, A.C, Frederick, C.A, Lippard, S.J, Nordlund, P. | | Deposit date: | 1994-02-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane.
Nature, 366, 1993
|
|
1MO5
 
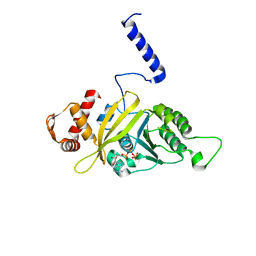 | |
3UL4
 
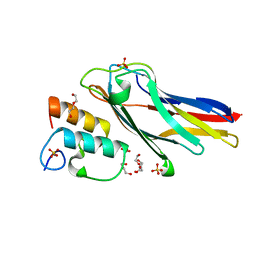 | | Crystal structure of Coh-OlpA(Cthe_3080)-Doc918(Cthe_0918) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome enzyme, dockerin type I, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
6QS5
 
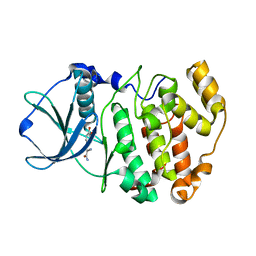 | |
1MPW
 
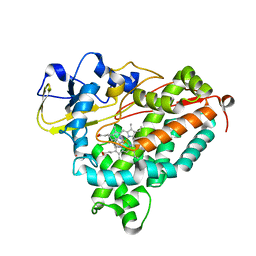 | | Molecular Recognition in (+)-a-Pinene Oxidation by Cytochrome P450cam | | Descriptor: | (+)-alpha-Pinene, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Bell, S.G, Chen, X, Sowden, R.J, Xu, F, Willams, J.N, Wong, L.-L, Rao, Z. | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam
J.Am.Chem.Soc., 125, 2003
|
|
6QQF
 
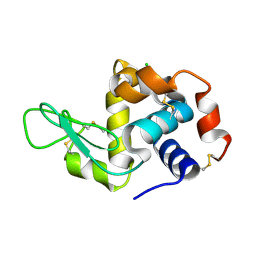 | |
3UM9
 
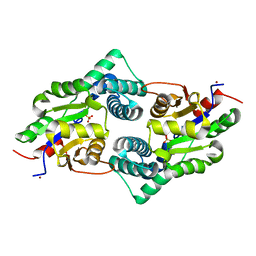 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
4OU9
 
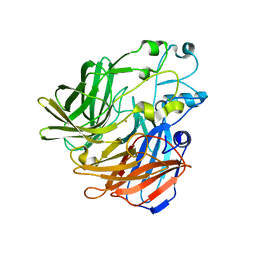 | | Crystal structure of apocarotenoid oxygenase in the presence of Triton X-100 | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Palczewski, K, Kiser, P.D. | | Deposit date: | 2014-02-15 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of Carotenoid Isomerase Activity in a Prototypical Carotenoid Cleavage Enzyme, Apocarotenoid Oxygenase (ACO).
J.Biol.Chem., 289, 2014
|
|
6QT2
 
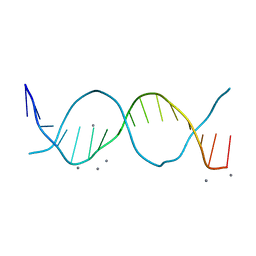 | | Radiation damage study on a 16mer DNA segment, structure at 6.2 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
1MO4
 
 | |
6QT6
 
 | | Radiation damage study on a 16mer DNA segment, structure at 29.2 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
1MPU
 
 | | Crystal Structure of the free human NKG2D immunoreceptor | | Descriptor: | NKG2-D type II integral membrane protein, PHOSPHATE ION | | Authors: | McFarland, B.J, Kortemme, T, Baker, D, Strong, R.K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry Recognizing Asymmetry: Analysis of the Interactions between the C-Type Lectin-like Immunoreceptor NKG2D and MHC
Class I-like Ligands
Structure, 11, 2003
|
|
3UXG
 
 | | Crystal structure of RFXANK | | Descriptor: | DNA-binding protein RFXANK, Histone deacetylase 4, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chao, X, Bian, C, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
7LOQ
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6QT1
 
 | | Radiation damage study on a 16mer DNA segment, structure at 0.48 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
6QT4
 
 | | Radiation damage study on a 16mer DNA segment, structure at 17.7 MGy dose | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Bugris, V, Harmat, V, Ferenc, G, Brockhauser, S, Carmichael, I, Garman, E.F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation-damage investigation of a DNA 16-mer.
J.Synchrotron Radiat., 26, 2019
|
|
6QU7
 
 | | Crystal structure of human DHODH in complex with BAY 2402234 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Friberg, A, Gradl, S. | | Deposit date: | 2019-02-26 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies.
Leukemia, 33, 2019
|
|
7RGE
 
 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | Descriptor: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
3V09
 
 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
7LN9
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LPL
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, dataset 3 (merged) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3UCC
 
 | | Asymmetric complex of human neuron specific enolase-1-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
7LPM
 
 | | X-ray radiation damage series on Lysozyme at 277K, multi-conformer model, crystal 2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LLP
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LN8
 
 | | X-ray radiation damage series on Lysozyme at 277K, crystal structure, dataset 3 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
