6PQX
 
 | | Cryo-EM structure of HzTransib/nicked TIR substrate DNA hairpin forming complex (HFC) | | Descriptor: | CALCIUM ION, DNA (5'-D(P*CP*AP*CP*GP*GP*TP*GP*GP*AP*TP*CP*GP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*GP*CP*CP*TP*AP*GP*AP*TP*CP*T)-3'), ... | | Authors: | Liu, C, Yang, Y, Schatz, D.G. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of a RAG-like transposase during cut-and-paste transposition.
Nature, 575, 2019
|
|
4YWM
 
 | |
2MRN
 
 | |
2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
4AUW
 
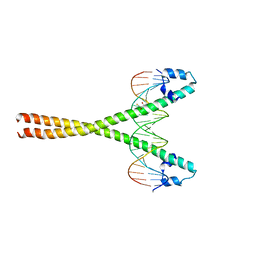 | | CRYSTAL STRUCTURE OF THE BZIP HOMODIMERIC MAFB IN COMPLEX WITH THE C- MARE BINDING SITE | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C-MARE BINDING SITE (5'-D(*AP*TP*AP*AP*TP*GP*CP*TP* GP*AP*CP*GP*TP*CP*AP*GP*CP*AP*AP*TP*T)-3'), MERCURY (II) ION, ... | | Authors: | Textor, L.C, Holton, S, Wilmanns, M. | | Deposit date: | 2012-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of the mouse transcription factor MafB in complex with its DNA-recognition motif Cmare
Acta Crystallogr Sect F Struct Biol Cryst Commun., 63, 2007
|
|
4YWK
 
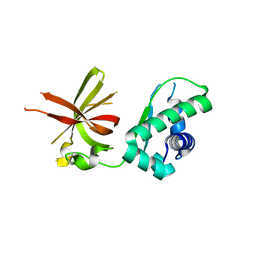 | |
6S58
 
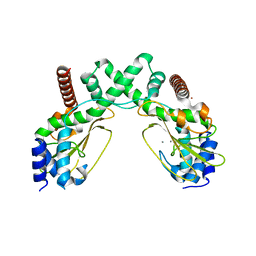 | | AvaII restriction endonuclease in the absence of nucleic acids | | Descriptor: | CALCIUM ION, Type II site-specific deoxyribonuclease, UNKNOWN ATOM OR ION | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
4YWL
 
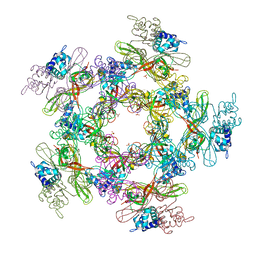 | |
7LO5
 
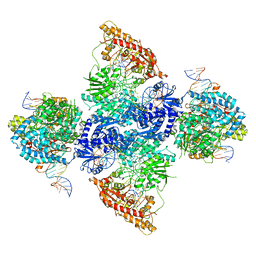 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
7LVV
 
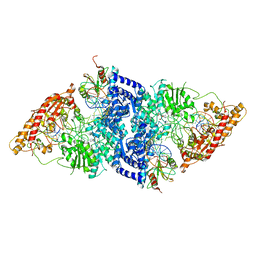 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
5A77
 
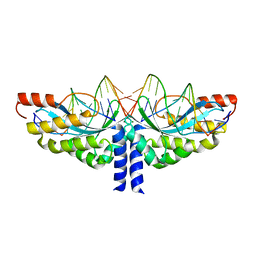 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA cleaved | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP* GP*AP*DGP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
6JGX
 
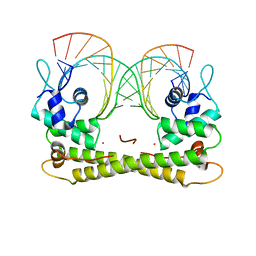 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2Q7U
 
 | |
8WD8
 
 | | Cryo-EM structure of TtdAgo-guide DNA-target DNA complex | | Descriptor: | Argonaute family protein, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
2ITJ
 
 | |
7Z0O
 
 | | Structure of transcription factor UAF in complex with TBP and 35S rRNA promoter DNA | | Descriptor: | Histone H3, Histone H4, Non-template DNA, ... | | Authors: | Baudin, F, Murciano, B, Fung, H.K.H, Fromm, S.A, Mueller, C.W. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of RNA polymerase I selection by transcription factor UAF.
Sci Adv, 8, 2022
|
|
1AOI
 
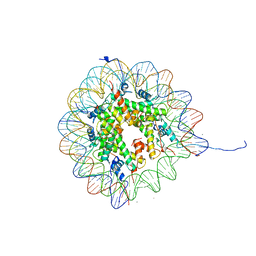 | | COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Luger, K, Maeder, A.W, Richmond, R.K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 1997-07-03 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the nucleosome core particle at 2.8 A resolution.
Nature, 389, 1997
|
|
1IXC
 
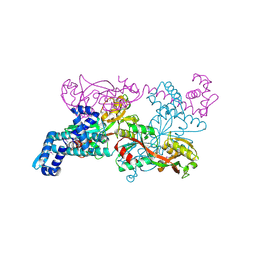 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IZ1
 
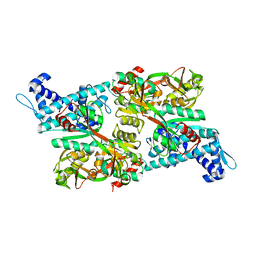 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
5K7H
 
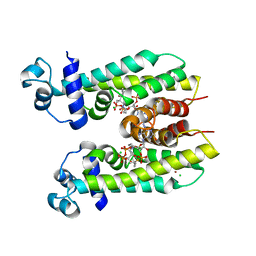 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | Descriptor: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
3F1W
 
 | |
5OHQ
 
 | |
2IPR
 
 | |
8FTJ
 
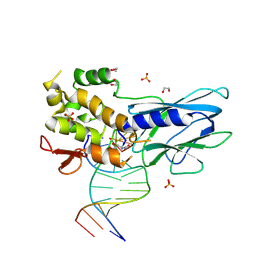 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
5K7F
 
 | | Crystal structure of apo AibR | | Descriptor: | ACETATE ION, Transcriptional regulator, TetR family | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
