4HDQ
 
 | |
4HDO
 
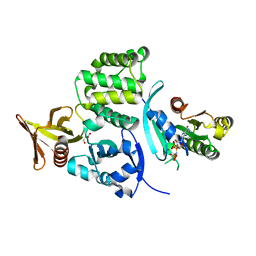 | | Crystal structure of the binary Complex of KRIT1 bound to the Rap1 GTPase | | Descriptor: | GLYCEROL, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2012-10-02 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) Bound to Both the Rap1 GTPase and the Heart of Glass (HEG1) Cytoplasmic Tail.
J.Biol.Chem., 288, 2013
|
|
4CW5
 
 | |
6RD6
 
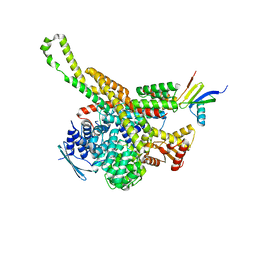 | | CryoEM structure of Polytomella F-ATP synthase, focussed refinement of upper peripheral stalk | | Descriptor: | ASA-2: Polytomella F-ATP synthase associated subunit 2, ATP synthase subunit alpha, Mitochondrial ATP synthase associated protein ASA4, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RD8
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 2, focussed refinement of Fo and peripheral stalk | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
1GT8
 
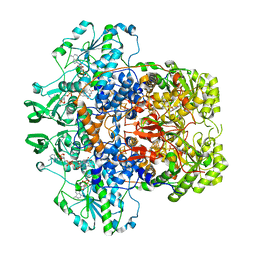 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND URACIL-4-ACETIC ACID | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
1GTE
 
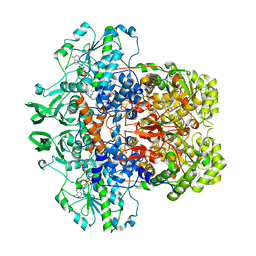 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY COMPLEX WITH 5-IODOURACIL | | Descriptor: | 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
6RD4
 
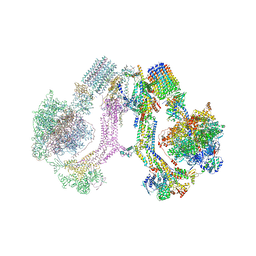 | | CryoEM structure of Polytomella F-ATP synthase, Full dimer, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RD7
 
 | | CryoEM structure of Polytomella F-ATP synthase, c-ring position 1, focussed refinement of Fo and peripheral stalk | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RD5
 
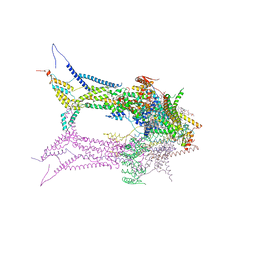 | | CryoEM structure of Polytomella F-ATP synthase, focussed refinement of Fo and peripheral stalk, C2 symmetry | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ATP synthase associated protein ASA1, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
4JTA
 
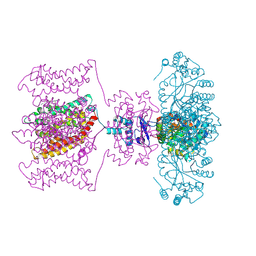 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | MacKinnon, R, Banerjee, A, Lee, A, Campbell, E. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
5X8U
 
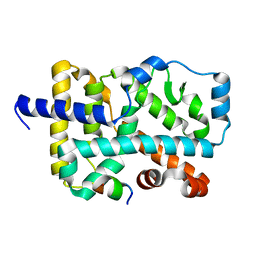 | | Crystal Structure of the wild Human ROR gamma Ligand Binding Domain. | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
5X8W
 
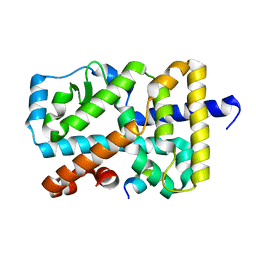 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain. | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
9B9H
 
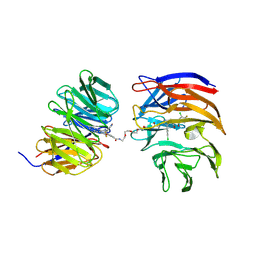 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40333 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(17E)-18-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-16-oxo-3,6,9,12-tetraoxa-15-azaoctadec-17-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Alvarez, H.G, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-02 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9B9W
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40792 | | Descriptor: | (4P)-N-[(1R)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-[(25E)-1-{(1P)-6-fluoro-3'-[4-fluoro-2-(trifluoromethyl)benzamido]-4'-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-1,24-dioxo-5,8,11,14,17,20-hexaoxa-2,23-diazahexacos-25-en-26-yl]-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Mamai, A, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9B9T
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-40407 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-[(17-{(4P)-4-[(2P)-4-{[(1R)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-1H-pyrazol-1-yl}-16-oxo-3,6,9,12-tetraoxa-15-azaheptadecan-1-yl)carbamoyl]-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Krausser, C, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9BA2
 
 | | Crystal structure of the binary complex of DCAF1 and WDR5 | | Descriptor: | DDB1- and CUL4-associated factor 1, IMIDAZOLE, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Srivastava, S, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
2WBL
 
 | | Three-dimensional structure of a binary ROP-PRONE complex | | Descriptor: | RAC-LIKE GTP-BINDING PROTEIN ARAC2, RHO OF PLANTS GUANINE NUCLEOTIDE EXCHANGE FACTOR 8 | | Authors: | Thomas, C, Fricke, I, Weyand, M, Berken, A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 3D Structure of a Binary Rop-Prone Complex: The Final Intermediate for a Complete Set of Molecular Snapshots of the Ropgef Reaction.
Biol.Chem., 390, 2009
|
|
9DLW
 
 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-41114 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(32E)-33-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-31-oxo-3,6,9,12,15,18,21,24,27-nonaoxa-30-azatritriacont-32-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Mamai, A, Wilson, B.J, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-09-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
3IAB
 
 | | Crystal structure of RNase P /RNase MRP proteins Pop6, Pop7 in a complex with the P3 domain of RNase MRP RNA | | Descriptor: | P3 domain of the RNA component of RNase MRP, Ribonucleases P/MRP protein subunit POP6, Ribonucleases P/MRP protein subunit POP7, ... | | Authors: | Perederina, A, Esakova, O.A, Quan, C, Khanova, E, Krasilnikov, A.S. | | Deposit date: | 2009-07-13 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain
Embo J., 29, 2010
|
|
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | Descriptor: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | Authors: | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
4R3N
 
 | | Crystal structure of the ternary complex of sp-ASADH with NADP and 1,2,3-Benzenetricarboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2014-08-16 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1GTH
 
 | | DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY COMPLEX WITH NADPH AND 5-IODOURACIL | | Descriptor: | (5S)-5-IODODIHYDRO-2,4(1H,3H)-PYRIMIDINEDIONE, 5-IODOURACIL, DIHYDROPYRIMIDINE DEHYDROGENASE, ... | | Authors: | Dobritzsch, D, Ricagno, S, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2002-01-15 | | Release date: | 2002-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the productive ternary complex of dihydropyrimidine dehydrogenase with NADPH and 5-iodouracil. Implications for mechanism of inhibition and electron transfer.
J. Biol. Chem., 277, 2002
|
|
7U1N
 
 | | Crystal structure of the Anopheles darlingi AD-118 long form D7 salivary protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-ETHOXYETHANOL, ... | | Authors: | Alvarenga, P.H, Gittis, A.G, Garboczi, D.N, Andersen, J.F. | | Deposit date: | 2022-02-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
3ORG
 
 | |
