7ASG
 
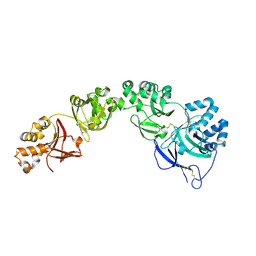 | | TGFBIp mutant R555W | | Descriptor: | Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Nielsen, N.S, Gadeberg, T.A.F, Andersen, G.R. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutation-induced dimerization of transforming growth factor-beta-induced protein may drive protein aggregation in granular corneal dystrophy.
J.Biol.Chem., 297, 2021
|
|
3DVF
 
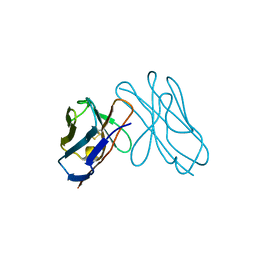 | |
4A7F
 
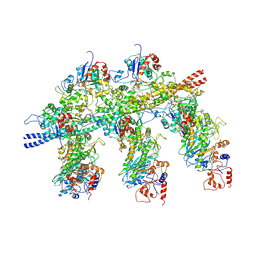 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
2WZ1
 
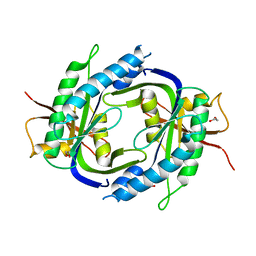 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN SOLUBLE GUANYLATE CYCLASE 1 BETA 3. | | Descriptor: | 1,2-ETHANEDIOL, GUANYLATE CYCLASE SOLUBLE SUBUNIT BETA-1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Muniz, J, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of the Catalytic Domain of Human Soluble Guanylate Cyclase.
Plos One, 8, 2013
|
|
4H6U
 
 | | Tubulin acetyltransferase mutant | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Roll-Mecak, A, Kizub, V, Szyk, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4482 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
2PRO
 
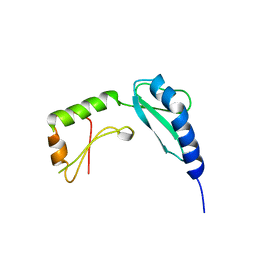 | |
4H6Z
 
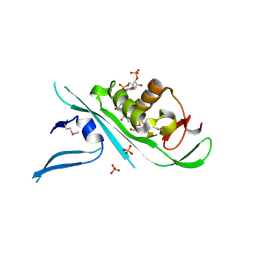 | | Tubulin acetyltransferase | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION | | Authors: | Kizub, L, Szyk, A, Piszczek, G, Roll-Mecak, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
2OBT
 
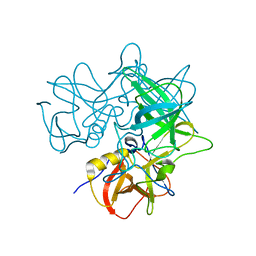 | |
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
1V3T
 
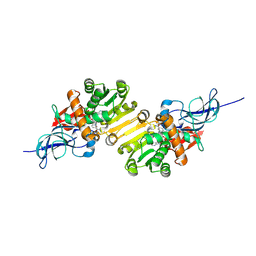 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
5DO2
 
 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3R2X
 
 | |
3G6D
 
 | | Crystal structure of the complex between CNTO607 Fab and IL-13 | | Descriptor: | CNTO607 Fab Heavy chain, CNTO607 Fab Light chain, Interleukin-13, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Epitope mapping of anti-interleukin-13 neutralizing antibody CNTO607.
J.Mol.Biol., 389, 2009
|
|
2OBS
 
 | |
4DIH
 
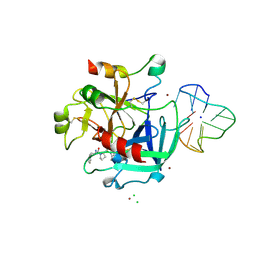 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of sodium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
4A7N
 
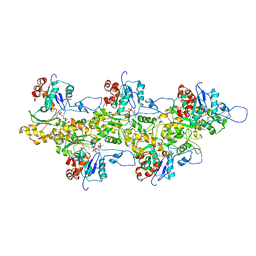 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
2OBR
 
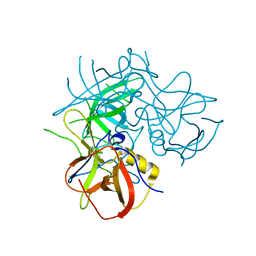 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
2OHO
 
 | |
2NRG
 
 | |
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
3EE7
 
 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
3HKB
 
 | | Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HKC
 
 | | Tubulin-ABT751: RB3 stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HKE
 
 | | Tubulin-T138067: RB3 stathmin-like domain complex | | Descriptor: | 2,3,4,5,6-pentafluoro-N-(3-fluoro-4-methoxyphenyl)benzenesulfonamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dorleans, A, Gigant, B, Ravelli, R.B.G, Mailliet, P, Mikol, V, Knossow, M. | | Deposit date: | 2009-05-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Variations in the colchicine-binding domain provide insight into the structural switch of tubulin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
