5NBD
 
 | | PglK flippase in complex with inhibitory nanobody | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nanobody, WlaB protein | | Authors: | Perez, C, Pardon, E, Steyaert, J, Locher, K.P. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody.
Sci Rep, 7, 2017
|
|
5OD5
 
 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | Authors: | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | Deposit date: | 2017-07-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
5MJI
 
 | | Crystal Structure of RosB with bound intermediate OHC-RP (8-demethyl-8-formylriboflavin-5'-phosphate) | | Descriptor: | BRAMP domain protein, [(2~{S},3~{R},4~{R})-5-[8-methanoyl-7-methyl-2,4-bis(oxidanylidene)-1~{H}-benzo[g]pteridin-10-ium-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7KKG
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
7KLS
 
 | |
7KLY
 
 | |
1CVJ
 
 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
4W5H
 
 | |
5BSW
 
 | |
5BSV
 
 | |
5BSM
 
 | |
5BST
 
 | |
5BSU
 
 | | Crystal structure of 4-coumarate:CoA ligase complexed with caffeoyl adenylate | | Descriptor: | 4-coumarate--CoA ligase 2, 5'-O-[(R)-{[(2E)-3-(3,4-dioxocyclohexa-1,5-dien-1-yl)prop-2-enoyl]oxy}(hydroxy)phosphoryl]adenosine, GLYCEROL, ... | | Authors: | Li, Z, Nair, S.K. | | Deposit date: | 2015-06-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Specificity and Flexibility in a Plant 4-Coumarate:CoA Ligase.
Structure, 23, 2015
|
|
4M0K
 
 | | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CALCIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775.
To be Published
|
|
5JJQ
 
 | | Crystal structure of IdnL1 | | Descriptor: | 5'-O-[(R)-{[(3S)-3-aminobutanoyl]oxy}(hydroxy)phosphoryl]adenosine, AMP-dependent synthetase and ligase, CHLORIDE ION | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2016-04-25 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical characterization and structural insight into aliphatic beta-amino acid adenylation enzymes IdnL1 and CmiS6
Proteins, 85, 2017
|
|
5JFF
 
 | | E. coli EcFicT mutant G55R in complex with EcFicA | | Descriptor: | CHLORIDE ION, Probable adenosine monophosphate-protein transferase fic, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Stanger, F.V, Schirmer, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Escherichia coli Fic Toxin-Like Protein in Complex with Its Cognate Antitoxin.
Plos One, 11, 2016
|
|
5JFZ
 
 | | E. coli EcFicT in complex with EcFicA mutant E28G | | Descriptor: | Probable adenosine monophosphate-protein transferase fic, Uncharacterized protein YhfG | | Authors: | Stanger, F.V, Schirmer, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Escherichia coli Fic Toxin-Like Protein in Complex with Its Cognate Antitoxin.
Plos One, 11, 2016
|
|
8Q2B
 
 | | E. coli Adenylate Kinase variant D158A (AK D158A) showing significant changes to the stacking of catalytic arginine side chains | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Sauer, U.H, Wolf-Watz, M, Nam, K. | | Deposit date: | 2023-08-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.
J.Chem.Inf.Model., 64, 2024
|
|
7TDJ
 
 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 1(Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDG
 
 | |
7TDK
 
 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 3 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDH
 
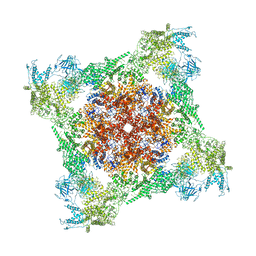 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in open conformation | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDI
 
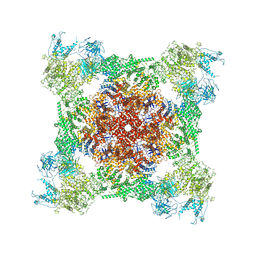 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 2 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
4MGI
 
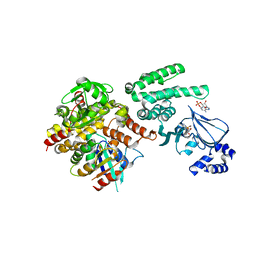 | | Selective activation of Epac1 and Epac2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
4MGK
 
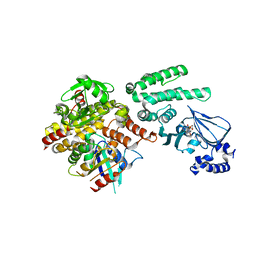 | | Selective activation of Epac1 and Epac2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
