5W1W
 
 | | Structure of the HLA-E-VMAPRTLVL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
9FNS
 
 | |
5W1V
 
 | | Structure of the HLA-E-VMAPRTLIL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
5U7F
 
 | |
5U7H
 
 | |
5UHY
 
 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
5UOY
 
 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 16j (6-(4-Methoxybenzyl)-9-((tetrahydro-2H-pyran-4-yl)methyl)-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-methoxyphenyl)methyl]-9-[(oxan-4-yl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
5UP0
 
 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 3 (6-(4-chlorobenzyl)-8,9,10,11-tetrahydrobenzo[4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-chlorophenyl)methyl]-8,9,10,11-tetrahydro[1]benzothieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
5L4O
 
 | |
2XSC
 
 | | Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli | | Descriptor: | SHIGA-LIKE TOXIN 1 SUBUNIT B, ZINC ION | | Authors: | Stein, P.E, Boodhoo, A, Tyrrell, G.J, Brunton, J.L, Oeffner, R.D, Bunkoczi, G, Read, R.J. | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of the Cell-Binding B Oligomer of Verotoxin-1 from E. Coli.
Nature, 355, 1992
|
|
1LTB
 
 | |
7BH8
 
 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
7UWH
 
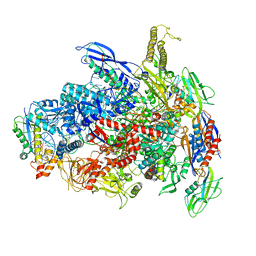 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate | | Descriptor: | DNA (59-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
7UWE
 
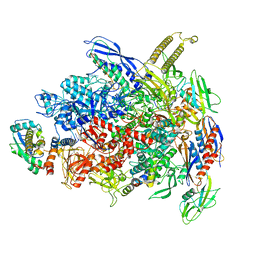 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
5W3Q
 
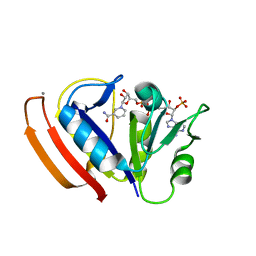 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
6HPB
 
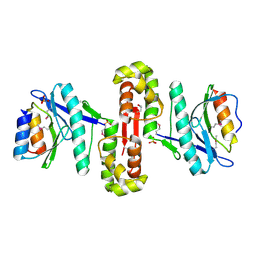 | |
3KXP
 
 | | Crystal Structure of E-2-(Acetamidomethylene)succinate Hydrolase | | Descriptor: | Alpha-(N-acetylaminomethylene)succinic acid hydrolase, CHLORIDE ION | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure determination and characterization of the vitamin B(6) degradative enzyme (E)-2-(acetamidomethylene)succinate hydrolase.
Biochemistry, 49, 2010
|
|
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
4ZT1
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in x-dimer conformation | | Descriptor: | CALCIUM ION, Cadherin-1 | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
6HPC
 
 | |
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
1MBB
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-3(N-ACETYLGLUCOSAMINYL)BUTYRIC ACID | | Authors: | Benson, T.E, Lees, W.J, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (E)-enolbutyryl-UDP-N-acetylglucosamine as a mechanistic probe of UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 35, 1996
|
|
1ED3
 
 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
5U7E
 
 | |
4ZSW
 
 | | Pig Brain GABA-AT inactivated by (E)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
