2GFI
 
 | | Crystal structure of the phytase from D. castellii at 2.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Hoh, F. | | Deposit date: | 2006-03-22 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Debaryomyces castellii CBS 2923 phytase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2VQ8
 
 | | RNASE ZF-1A | | Descriptor: | CHLORIDE ION, RNASE ZF-1A | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications.
J.Mol.Biol., 380, 2008
|
|
2VQ9
 
 | | RNASE ZF-3E | | Descriptor: | CHLORIDE ION, RNASE 1 | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications
J.Mol.Biol., 380, 2008
|
|
1KL9
 
 | |
1GX3
 
 | | M. smegmatis arylamine N-acetyl transferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Mushtaq, A, Kawamura, A, Sinclair, J, Sim, E, Noble, M. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Arylamine N-Acetyltransferase from Mycobacterium Smegmatis-an Enzyme which Inactivates the Anti-Tubercular Drug, Isoniazid
J.Mol.Biol., 318, 2002
|
|
2PQT
 
 | | Human N-acetyltransferase 1 | | Descriptor: | Arylamine N-acetyltransferase 1, CHLORIDE ION, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Grant, D.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis of Substrate-binding Specificity of Human Arylamine N-Acetyltransferases.
J.Biol.Chem., 282, 2007
|
|
1HDH
 
 | | Arylsulfatase from Pseudomonas aeruginosa | | Descriptor: | Arylsulfatase, CALCIUM ION, SULFATE ION | | Authors: | Boltes, I, Czapinska, H, Kahnert, A, von Buelow, R, Dirks, T, Schmidt, B, von Figura, K, Kertesz, M.A, Uson, I. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 A Structure of Arylsulfatase from Pseudomonas Aeruginosa Establishes the Catalytic Mechanism of Sulfate Ester Cleavage in the Sulfatase Family.
Structure, 9, 2001
|
|
4BGF
 
 | | The 3D-structure of arylamine-N-acetyltransferase from M. tuberculosis | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Lowe, E.D, McDonough, M.A, Shaw Stewart, P.D, Kolek, S.A, Sim, E, Garman, E.F. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2G2K
 
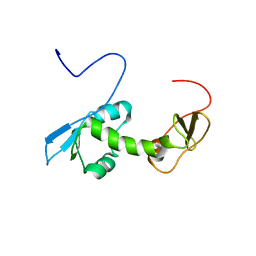 | | NMR structure of an N-terminal fragment of the eukaryotic initiation factor 5 (eIF5) | | Descriptor: | Eukaryotic translation initiation factor 5 | | Authors: | Conte, M.R, Kelly, G, Babon, J, Sanfelice, D, Smerdon, S.J, Proud, C.G. | | Deposit date: | 2006-02-16 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors
Biochemistry, 45, 2006
|
|
2J76
 
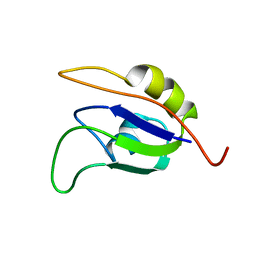 | | Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4B | | Authors: | Fleming, K, Ghuman, J, Yuan, X.M, Simpson, P, Szendroi, A, Matthews, S, Curry, S. | | Deposit date: | 2006-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and RNA Interactions of the RNA Recognition Motif from Eukaryotic Translation Initiation Factor 4B.
Biochemistry, 42, 2003
|
|
1E2T
 
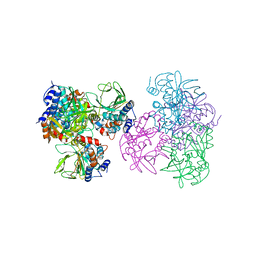 | | Arylamine N-acetyltransferase (NAT) from Salmonella typhimurium | | Descriptor: | N-HYDROXYARYLAMINE O-ACETYLTRANSFERASE | | Authors: | Sinclair, J.C, Sandy, J, Delgoda, R, Sim, E, Noble, M.E.M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Nat.Struct.Biol., 7, 2000
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
1EE5
 
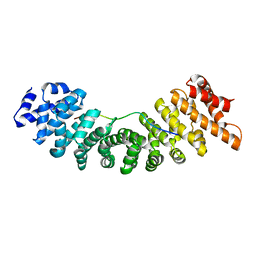 | |
1BK6
 
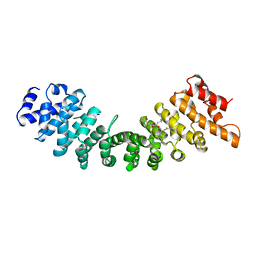 | | KARYOPHERIN ALPHA (YEAST) + SV40 T ANTIGEN NLS | | Descriptor: | KARYOPHERIN ALPHA, LARGE T ANTIGEN | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1EE4
 
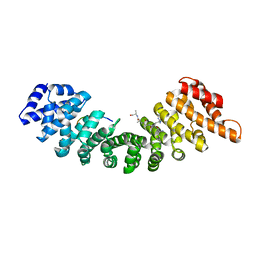 | |
1TXT
 
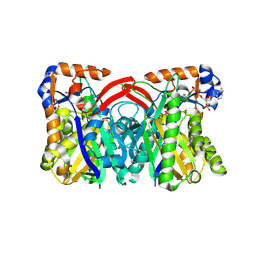 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
2CRD
 
 | | ANALYSIS OF SIDE-CHAIN ORGANIZATION ON A REFINED MODEL OF CHARYBDOTOXIN: STRUCTURAL AND FUNCTIONAL IMPLICATIONS | | Descriptor: | CHARYBDOTOXIN | | Authors: | Bontems, F, Roumestand, C, Gilquin, B, Menez, A, Toma, F. | | Deposit date: | 1993-02-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications.
Biochemistry, 31, 1992
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
3F7K
 
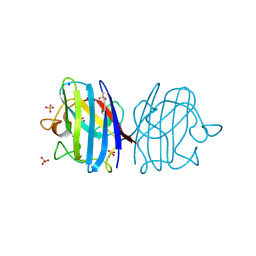 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
3F7L
 
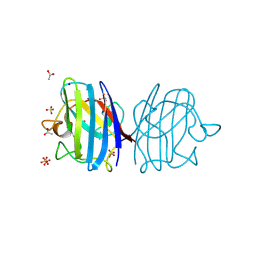 | | X-ray Crystal Structure of Alvinella pompejana Cu,Zn Superoxide Dismutase | | Descriptor: | ACETIC ACID, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
8P2K
 
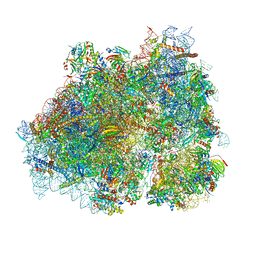 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
3S8R
 
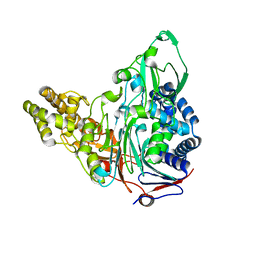 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
3MP5
 
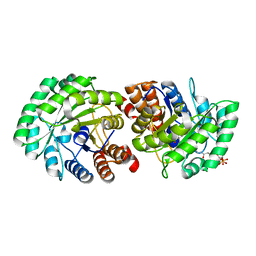 | | Crystal Structure of Human Lyase R41M in complex with HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, Hydroxymethylglutaryl-CoA lyase, MAGNESIUM ION | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3HB5
 
 | | Binary and ternary crystal structures of a novel inhibitor of 17 beta-HSD type 1: a lead compound for breast cancer therapy | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazumdar, M, Lin, S.-X. | | Deposit date: | 2009-05-04 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binary and ternary crystal structure analyses of a novel inhibitor with 17beta-HSD type 1: a lead compound for breast cancer therapy.
Biochem.J., 424, 2009
|
|
5XAK
 
 | |
