5COR
 
 | |
5CMD
 
 | |
2L4N
 
 | |
2MGS
 
 | | Solution structure of CXCL5 | | Descriptor: | C-X-C motif chemokine 5 | | Authors: | Sepuru, K, Rajarathnam, K. | | Deposit date: | 2013-11-04 | | Release date: | 2014-04-02 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of CXCL5 - A Novel Chemokine and Adipokine Implicated in Inflammation and Obesity.
Plos One, 9, 2014
|
|
2N54
 
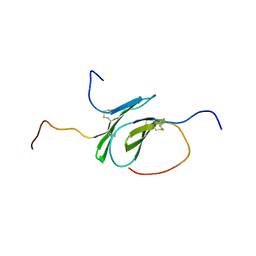 | | Solution structure of a disulfide stabilized XCL1 dimer | | Descriptor: | Lymphotactin | | Authors: | Tyler, R.C, Tuinstra, R.L, Peterson, F.F, Volkman, B.F. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering Metamorphic Chemokine Lymphotactin/XCL1 into the GAG-Binding, HIV-Inhibitory Dimer Conformation.
Acs Chem.Biol., 10, 2015
|
|
2MPM
 
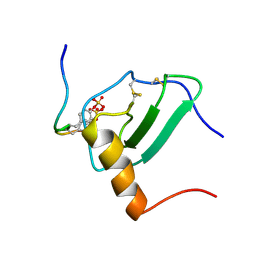 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
2NWG
 
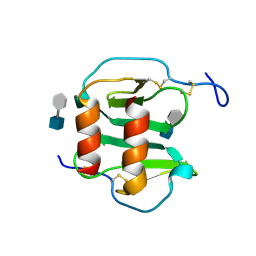 | | Structure of CXCL12:heparin disaccharide complex | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Stromal cell-derived factor 1 | | Authors: | Murphy, J.W, Cho, Y, Lolis, E. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and Functional Basis of CXCL12 (Stromal Cell-derived Factor-1{alpha}) Binding to Heparin
J.Biol.Chem., 282, 2007
|
|
2Q8T
 
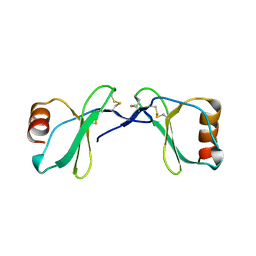 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
2Q8R
 
 | |
2MP1
 
 | |
1SDF
 
 | | SOLUTION STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
1U4L
 
 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4M
 
 | | human RANTES complexed to heparin-derived disaccharide III-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4P
 
 | | Crystal Structure of human RANTES mutant K45E | | Descriptor: | ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1RTN
 
 | |
1RHP
 
 | |
1RJT
 
 | |
1RTO
 
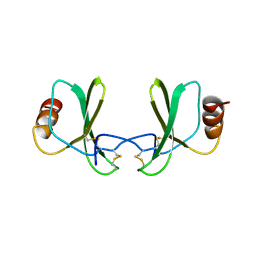 | |
1ROD
 
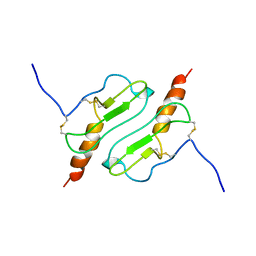 | | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN, NMR | | Descriptor: | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN | | Authors: | Roesch, P, Sticht, H, Auer, M, Schmitt, B, Besemer, J, Horcher, M, Kirsch, T, Lindley, I.J.D. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and activity of a chimeric interleukin-8-melanoma-growth-stimulatory-activity protein.
Eur.J.Biochem., 235, 1996
|
|
1TVX
 
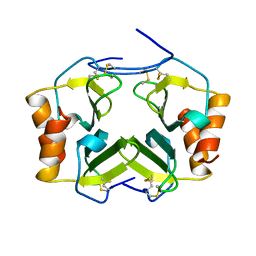 | |
1U4R
 
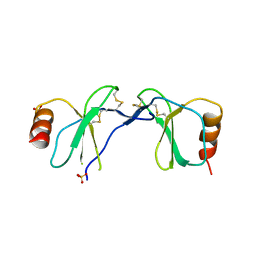 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VMP
 
 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
1MSH
 
 | |
1NCV
 
 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
