4YWK
 
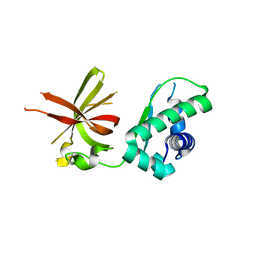 | |
6CJ5
 
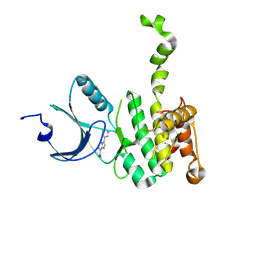 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-3-yl)imidazo[1,2-a]pyridine-8-carboxamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CKI
 
 | | Co-crystal structure of MNK2 in Complex With Inhibitor | | Descriptor: | 3,3-dimethyl-6-[(pyrimidin-4-yl)amino]-2,3-dihydroimidazo[1,5-a]pyridine-1,5-dione, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
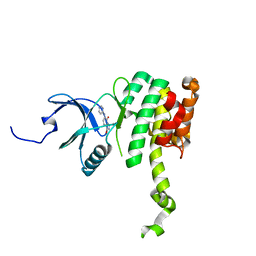
6CJH
 
 | | Co-crystal structure of MNK2 in complex with an inhibitor | | Descriptor: | 3-phenyl-5-(pyridin-4-yl)-1H-indazole, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
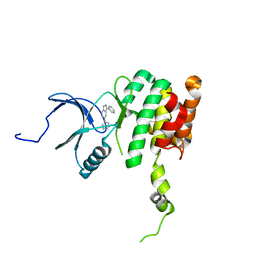
7BL1
 
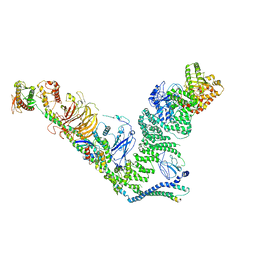 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
8JBT
 
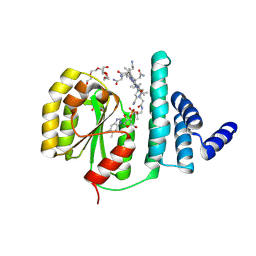 | | B12-binding domain from Chloracidobacterium thermophilum MerR family protein, anaerobic light state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALAMIN, GLYCEROL, ... | | Authors: | Zhang, S, Yu, Y. | | Deposit date: | 2023-05-09 | | Release date: | 2024-11-13 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SignatureFinder enables sequence mining to identify cobalamin-dependent photoreceptor proteins.
Febs J., 292, 2025
|
|
4MFZ
 
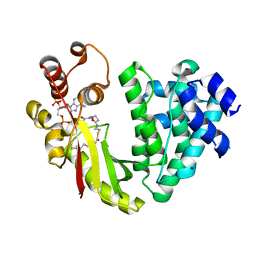 | | The crystal structure of acyltransferase in complex with decanoyl-CoA | | Descriptor: | Dbv8 protein, SODIUM ION, SULFATE ION, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFP
 
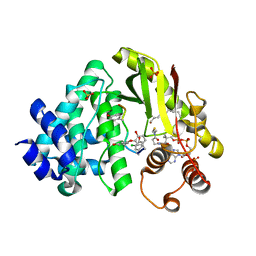 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFJ
 
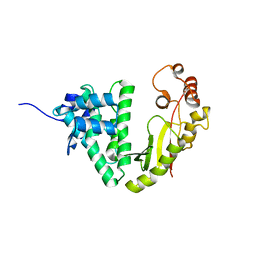 | | The crystal structure of acyltransferase | | Descriptor: | Putative uncharacterized protein tcp24 | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFL
 
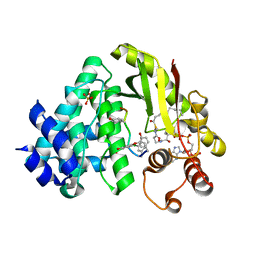 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and Tei pseudoaglycone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein tcp24, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4MFQ
 
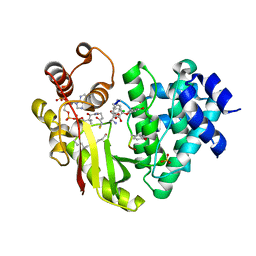 | | The crystal structure of acyltransferase in complex with CoA and 10C-Teicoplanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
6WPA
 
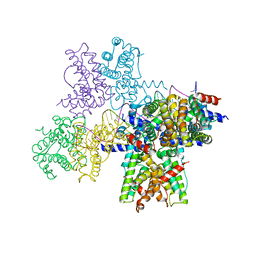 | |
6WP9
 
 | | AvaR1 bound to Avenolide | | Descriptor: | (5S)-5-[(6R)-6-hydroxy-6-methyl-5-oxooctyl]furan-2(5H)-one, AvaR1 | | Authors: | Kapoor, I, Olivares, P.J, Nair, S.K. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical basis for the regulation of biosynthesis of antiparasitics by bacterial hormones.
Elife, 9, 2020
|
|
9LFT
 
 | | Biochemical and structural characterization of a novel 4-hydroxyphenylacetate-3-monooxygenase from Geobacillus mahadii Geo-05 | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 4-Hydroxyphenylacetate-3-monooxygenase | | Authors: | Che Husain, N.A, Jamaluddin, H, Jonet, M.A, Padzil, A.M, Taib, A.Z.M, Zain, N.C, Bakar, M.F.A, Acharya, K.R, Gregory, K.S. | | Deposit date: | 2025-01-09 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural insights into GMHpaB: A thermostable 4-Hydroxyphenylacetate-3-monooxygenase with dual cofactor versatility.
Int.J.Biol.Macromol., 321, 2025
|
|
7L1X
 
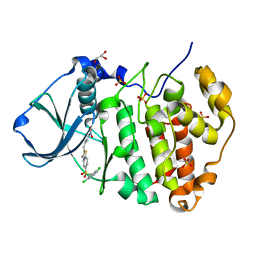 | | Structure of human CK2 alpha kinase (catalytic subunit) with the inhibitor 108600. | | Descriptor: | (2~{Z})-6-[[2,6-bis(chloranyl)phenyl]methylsulfonyl]-2-[[4-oxidanyl-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methylidene]-4~{H}-1,4-benzothiazin-3-one, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous CK2/TNIK/DYRK1 inhibition by 108600 suppresses triple negative breast cancer stem cells and chemotherapy-resistant disease.
Nat Commun, 12, 2021
|
|
4WWF
 
 | | High-resolution structure of two Ni-bound forms of the M123C mutant of C. metallidurans CnrXs | | Descriptor: | NICKEL (II) ION, Nickel and cobalt resistance protein CnrR, SODIUM ION | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kinnemann, S, Grosse, C, Schleuder, G, Petit-Hurtlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
4WWD
 
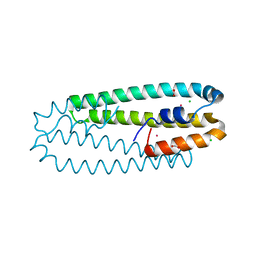 | | High-resolution structure of the Co-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
4WWB
 
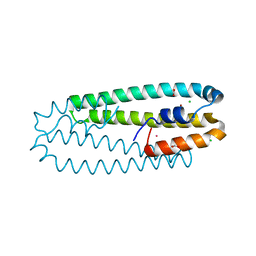 | | High-resolution structure of the Ni-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
6C6H
 
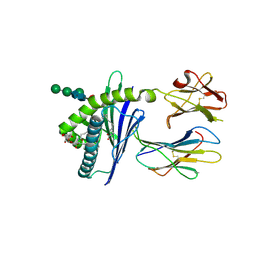 | | Structure of glycolipid aGSA[8,P5m] in complex with mouse CD1d | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(3-pentylphenyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)te trahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7X6H
 
 | |
7X6F
 
 | | Outer membrane lipoprotein QseG of Escherichia coli O157:H7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CITRATE ANION, ... | | Authors: | Matsumoto, K, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-03-07 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7X6G
 
 | |
5W43
 
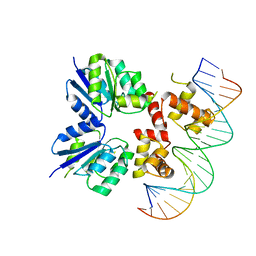 | | Structure of the two-component response regulator RcsB-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*TP*C)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Warwzak, Z, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
6YCF
 
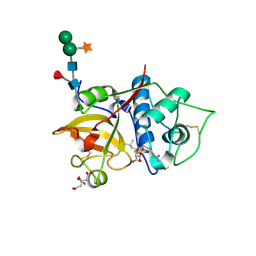 | | Structure the bromelain protease from Ananas comosus in complex with the E64 inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, FBSB, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCE
 
 | | Structure the bromelain protease from Ananas comosus with a thiomethylated active cysteine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
