6FC5
 
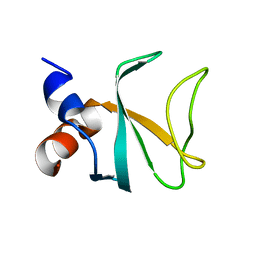 | | Bik1 CAP-Gly domain | | Descriptor: | Microtubule-associated protein | | Authors: | Kumar, A, Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Function Relationship of the Bik1-Bim1 Complex.
Structure, 26, 2018
|
|
6FC6
 
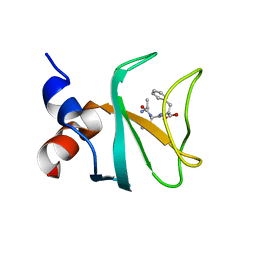 | |
7Q8V
 
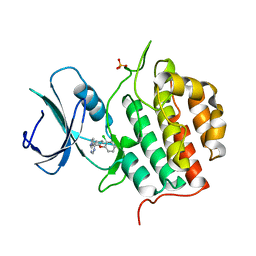 | | Crystal structure of TTBK1 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 1, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
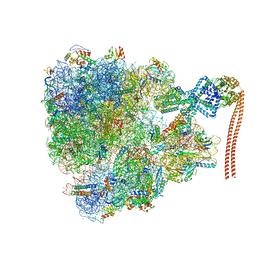
7QV3
 
 | |
7Q8W
 
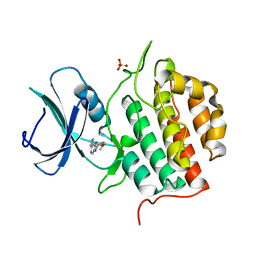 | | Crystal structure of TTBK1 in complex with VNG1.35 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Tau-tubulin kinase 1, ... | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7QV1
 
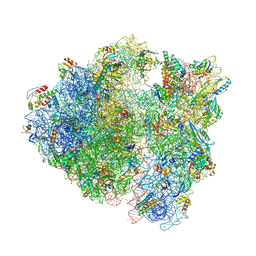 | | Bacillus subtilis collided disome (Leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QH4
 
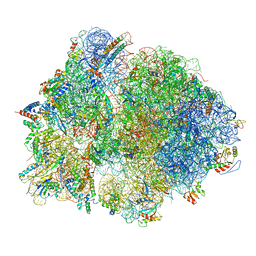 | | Structure of the B. subtilis disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.45 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QV2
 
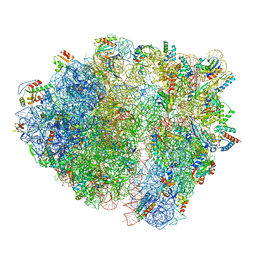 | | Bacillus subtilis collided disome (Collided 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QGU
 
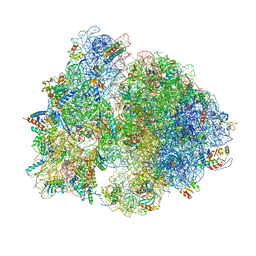 | | Structure of the B. subtilis disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QHW
 
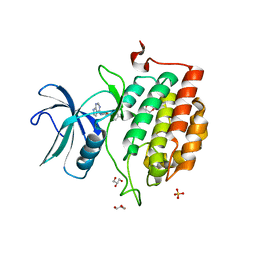 | | TTBK1 kinase domain in complex with inhibitor 29 | | Descriptor: | GLYCEROL, SULFATE ION, Tau-tubulin kinase 1, ... | | Authors: | Nozal, V, Liehta, D. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
2VDL
 
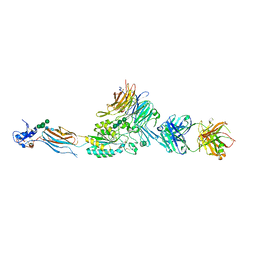 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
2ATS
 
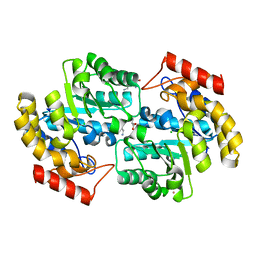 | | Dihydrodipicolinate synthase co-crystallised with (S)-lysine | | Descriptor: | CHLORIDE ION, D-LYSINE, POTASSIUM ION, ... | | Authors: | Devenish, S.R.A, Dobson, R.C.J, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-crystallisation of (S)-lysine-bound dihydrodipicolinate synthase from E. coli indicates that domain movements are not responsible for (S)-lysine inhibition
To be published
|
|
3X3F
 
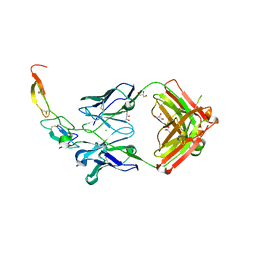 | |
4EOU
 
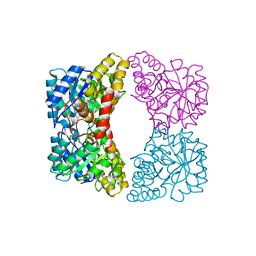 | |
7YF5
 
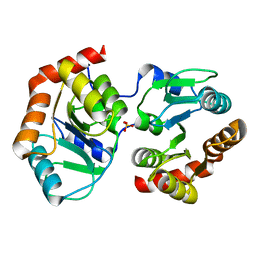 | |
7YAN
 
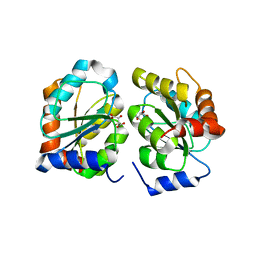 | |
4G07
 
 | |
4G09
 
 | |
7ZHN
 
 | | Crystal structure of TTBK1 in complex with AMG28 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-amino-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-11-yl)-2-methylbut-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHQ
 
 | | Crystal structure of TTBK1 in complex with compound 10 (7-009) | | Descriptor: | (3~{S})-1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-methyl-pent-1-yn-3-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHO
 
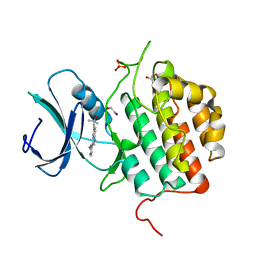 | | Crystal structure of TTBK1 in complex with compound 3 (7-001) | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(2-azanylpyrimidin-4-yl)-1~{H}-indol-5-yl]-2-methyl-but-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHP
 
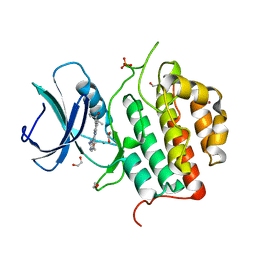 | | Crystal structure of TTBK1 in complex with compound 9 (7-005) | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-ethyl-pent-1-yn-3-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
4I9X
 
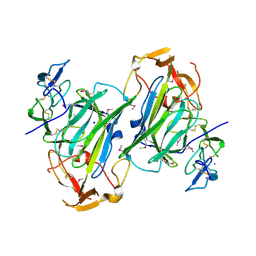 | |
8BW4
 
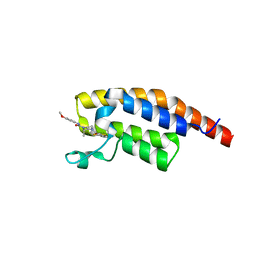 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
