5AFW
 
 | | Assembly of methylated LSD1 and CHD1 drives AR-dependent transcription and translocation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 1, ... | | Authors: | Metzger, E, Willmann, D, McMillan, J, Petroll, K, Metzger, P, Gerhardt, S, vonMaessenhausen, A, Schott, A.K, Espejo, A, Eberlin, A, Wohlwend, D, Schuele, K.M, Schleicher, M, Perner, S, Bedford, M.T, Dengjel, J, Flaig, R, Einsle, O, Schuele, R. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Assembly of Methylated Kdm1A and Chd1 Drives Androgen Receptor-Dependent Transcription and Translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3V4A
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 2 | | Descriptor: | (5R)-3-(3,4-dichlorophenyl)-5-(4-hydroxyphenyl)-1,5-dimethyl-2-thioxoimidazolidin-4-one, Androgen receptor, SULFATE ION | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
3V49
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 1 | | Descriptor: | 4-[(4R)-4-(4-hydroxyphenyl)-3,4-dimethyl-2,5-dioxoimidazolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, activator peptide, ... | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.-M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
5C9K
 
 | |
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7PLE
 
 | | ArsH of Paracoccus denitrificans | | Descriptor: | NADPH-dependent FMN reductase | | Authors: | Kryl, M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The ArsH Protein Product of the Paracoccus denitrificans ars Operon Has an Activity of Organoarsenic Reductase and Is Regulated by a Redox-Responsive Repressor.
Antioxidants, 11, 2022
|
|
6R1Q
 
 | | murine Neuroglobin under 2 kBar of argon | | Descriptor: | ARGON, CHLORIDE ION, Neuroglobin, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2019-03-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Argon-labelling protein crystals by high pressure cooling at 2000 Bar
To Be Published
|
|
6QAR
 
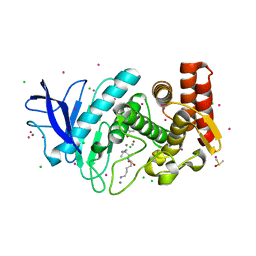 | |
1C66
 
 | |
1C6F
 
 | |
1C6I
 
 | |
1C6C
 
 | |
6RA8
 
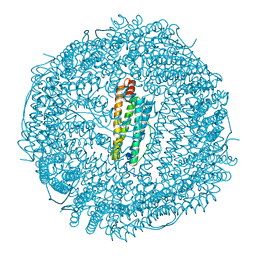 | |
6Q8S
 
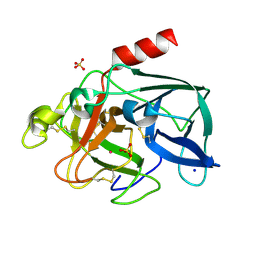 | | Elastase (PPE) under 2 kbar of argon | | Descriptor: | ARGON, Chymotrypsin-like elastase family member 1, SODIUM ION, ... | | Authors: | Prange, T, Carpentier, P. | | Deposit date: | 2018-12-16 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Labelling protein crystals by argon at 2 kbar
To be published
|
|
6Q8Q
 
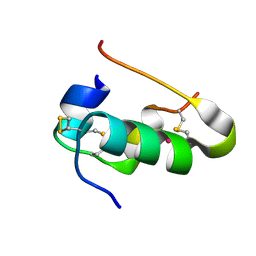 | |
5NVI
 
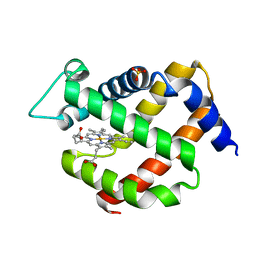 | |
2IA5
 
 | | T4 polynucleotide kinase/phosphatase with bound sulfate and magnesium. | | Descriptor: | ARSENIC, MAGNESIUM ION, Polynucleotide kinase, ... | | Authors: | Zhu, H, Smith, P.C, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2006-09-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis of the 3' phosphatase component of T4 polynucleotide kinase/phosphatase.
Virology, 366, 2007
|
|
1NLX
 
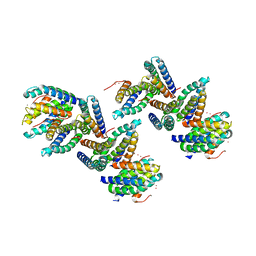 | | Crystal Structure of PHL P 6, A Major Timothy Grass Pollen Allergen Co-Crystallized with Zinc | | Descriptor: | ARSENIC, Pollen allergen Phl p 6, ZINC ION | | Authors: | Fedorov, A.A, Ball, T, Fedorov, E.V, Vrtala, S, Valenta, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure oh Phl p 6, a major timothy grass pollen allergen co-crystallized with Zinc
To be Published
|
|
1F0J
 
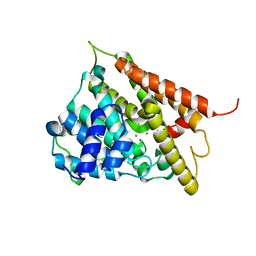 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | Descriptor: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | Authors: | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
5D8Q
 
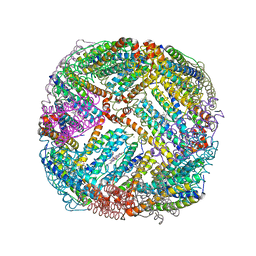 | | 2.20A resolution structure of BfrB (L68A) from Pseudomonas aeruginosa | | Descriptor: | ARSENIC, Ferroxidase, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
