3WEW
 
 | |
4YUC
 
 | | Crystal Structure of CorB derivatized with S-(2-acetamidoethyl) 4-methyl-3-oxohexanethioate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CorB, SODIUM ION | | Authors: | Zocher, G, Vilstrup, J, Stehle, T. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis of head to head polyketide fusion by CorB.
Chem Sci, 6, 2015
|
|
6UV5
 
 | |
6UI9
 
 | |
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
4Q1K
 
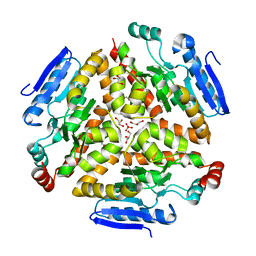 | |
4Q1J
 
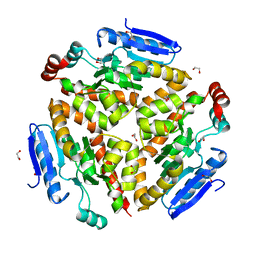 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1I
 
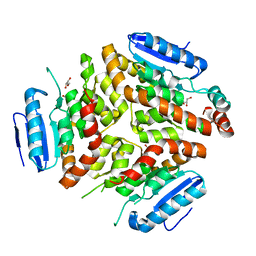 | |
1QR0
 
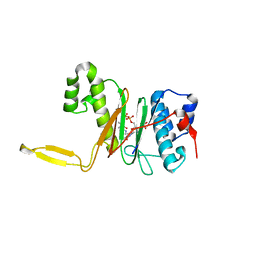 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
3CCF
 
 | |
5C1J
 
 | |
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
2B3M
 
 | | Crystal structure of protein AF1124 from Archaeoglobus fulgidus | | Descriptor: | hypothetical protein AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of protein AF1124 from Archaeoglobus fulgidus
To be Published
|
|
8HZ4
 
 | |
4Q1G
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
6A9N
 
 | |
1IQ6
 
 | | (R)-HYDRATASE FROM A. CAVIAE INVOLVED IN PHA BIOSYNTHESIS | | Descriptor: | (R)-SPECIFIC ENOYL-COA HYDRATASE, ISOPROPYL ALCOHOL | | Authors: | Hisano, T, Tsuge, T, Fukui, T, Iwata, T, Doi, Y. | | Deposit date: | 2001-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate
biosynthesis
J.Biol.Chem., 278, 2003
|
|
6MTW
 
 | | Lysosomal Phospholipase A2 in complex with Zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Group XV phospholipase A2, ... | | Authors: | Bouley, R, Tesmer, J.J.G. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural Basis of Lysosomal Phospholipase A2Inhibition by Zn2.
Biochemistry, 58, 2019
|
|
3R87
 
 | |
4Z0L
 
 | | The murine cyclooxygenase-2 complexed with a nido-dicarbaborate-containing indomethacin derivative | | Descriptor: | (R)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, (S)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Neumann, W, Banerjee, S, Hey-Hawkins, E, Marnett, L.J. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | nido-Dicarbaborate Induces Potent and Selective Inhibition of Cyclooxygenase-2.
Chemmedchem, 11, 2016
|
|
6BL3
 
 | | Crystal Complex of Cyclooxygenase-2 with indomethacin-butyldiamine-dansyl conjugate | | Descriptor: | 2-[1-(4-chlorobenzene-1-carbonyl)-5-methoxy-2-methyl-1H-indol-3-yl]-N-[4-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)butyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site.
J.Biol.Chem., 294, 2019
|
|
6BL4
 
 | | Crystal Complex of Cyclooxygenase-2 with indomethacin-ethylenediamine-dansyl conjugate | | Descriptor: | 2-[1-(4-chlorobenzene-1-carbonyl)-5-methoxy-2-methyl-1H-indol-3-yl]-N-[2-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)ethyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site.
J.Biol.Chem., 294, 2019
|
|
6QWU
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 5.5 with Mn2+ and CoA. | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QXQ
 
 | |
7VYU
 
 | |
