6LSF
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6RA/C2S6RB form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Wang, M, Shu, B, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
6LSH
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C2S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
8WA1
 
 | | The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2 | | Descriptor: | DNA (24-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8WA0
 
 | |
6LSG
 
 | | Crystal structure of the enterovirus 71 polymerase elongation complex (C0S6M form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Li, R, Wang, M, Jing, X, Gong, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures.
Nat Commun, 11, 2020
|
|
5F8I
 
 | | Enterovirus 71 Polymerase Elongation Complex (C1S2/3 Form) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, Genome polyprotein, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F8M
 
 | | Enterovirus 71 Polymerase Elongation Complex (C3S4/5 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F8G
 
 | | Enterovirus 71 Polymerase Elongation Complex (C1S1 Form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F8N
 
 | | Enterovirus 71 Polymerase Elongation Complex (C3S6 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, PYROPHOSPHATE 2-, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F8L
 
 | | Enterovirus 71 Polymerase Elongation Complex (C3S1 Form) | | Descriptor: | Genome polyprotein, RNA (35-MER), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.812 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Y6Z
 
 | | Crystal structure of the coxsackievirus A16 polymerase elongation complex | | Descriptor: | GLYCEROL, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Bi, P, Shu, B, Gong, P. | | Deposit date: | 2017-08-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the coxsackievirus A16 RNA-dependent RNA polymerase elongation complex reveals novel features in motif A dynamics
Virol Sin, 32, 2017
|
|
5F8H
 
 | | Enterovirus 71 Polymerase Elongation Complex (C1S1/2 Form) | | Descriptor: | Genome polyprotein, MAGNESIUM ION, RNA (35-MER), ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F8J
 
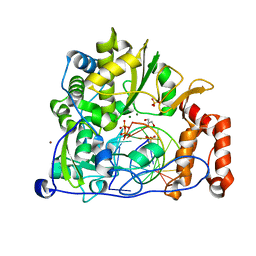 | | Enterovirus 71 Polymerase Elongation Complex (C1S4 Form) | | Descriptor: | GLYCEROL, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XLQ
 
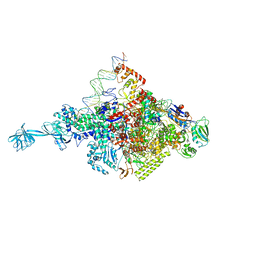 | |
4XLP
 
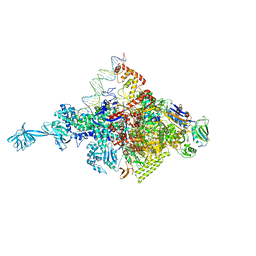 | |
4Y2C
 
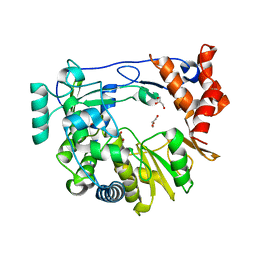 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
6L6V
 
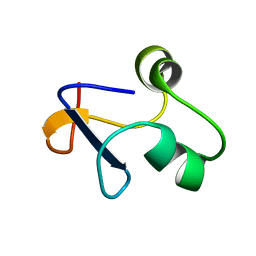 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
8QN8
 
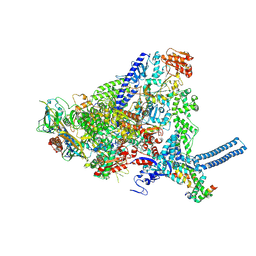 | | Mycobacterium smegmatis RNA polymerase in complex with HelD, SigA and RbpA in State II | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8QU6
 
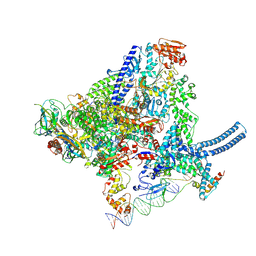 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD and an upstream-fork promoter fragment | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8EOE
 
 | |
8EHQ
 
 | |
8R6P
 
 | | Mycobacterium smegnatis RNA polymerase RP2-like transcription initiation complex with SigmaA, RbpA, HelD N-terminal domain and open promoter DNA | | Descriptor: | DNA 50-MER, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8R6R
 
 | | Mycobacterium smegnatis RNA polymerase RP2-like transcription initiation complex with SigmaA, RbpA and open promoter DNA | | Descriptor: | DNA 50-MER, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8R3M
 
 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD N-terminal, CO and PCh loop domain, and an upstream-fork promoter fragment; State III conformation | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-09 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
8R2M
 
 | | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD N-terminal domain and an upstream-fork promoter fragment; State III conformation | | Descriptor: | DNA 22-MER, DNA 31-MER, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Koval, T, Krasny, L, Dohnalek, J, Kouba, T. | | Deposit date: | 2023-11-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular insights into RNA polymerase recycling by mycobacterial HelD
To Be Published
|
|
