8YKY
 
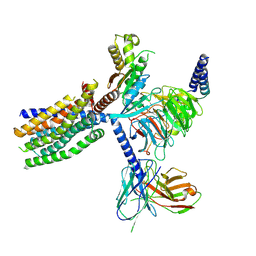 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQN
 
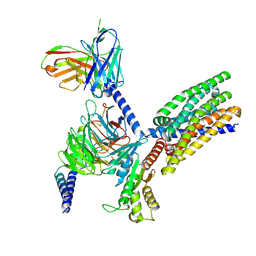 | | Structure of human class T GPCR TAS2R14-DNGi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQL
 
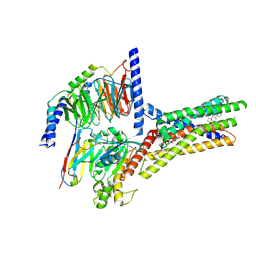 | | Structure of human class T GPCR TAS2R14-miniGs/gust complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQT
 
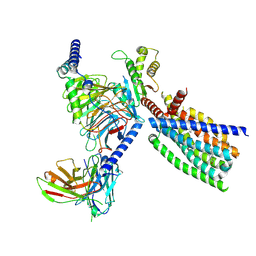 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQR
 
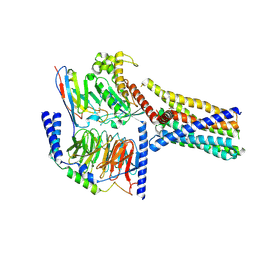 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8W9B
 
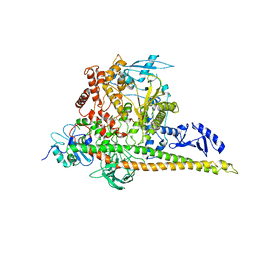 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | Descriptor: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
5TUV
 
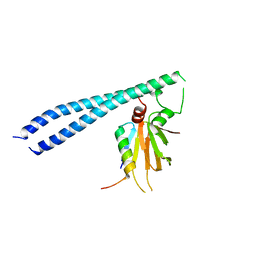 | |
5UL1
 
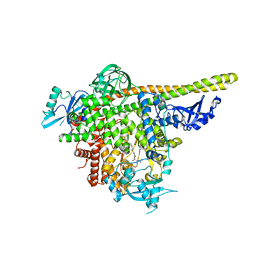 | | The co-structure of 3-amino-6-(4-((1-(dimethylamino)propan-2-yl)sulfonyl)phenyl)-N-phenylpyrazine-2-carboxamide and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | 3-amino-6-(4-{[(2S)-1-(dimethylamino)propan-2-yl]sulfonyl}phenyl)-N-phenylpyrazine-2-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Mamo, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
5UKJ
 
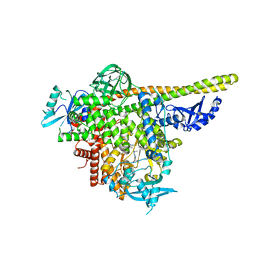 | | The co-structure of N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3- b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5- a]pyrazin-3-yl]benzenesulfonamide and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | N,N-dimethyl-4-[(6R)-6-methyl-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-3-yl]benzenesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Mamo, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
5UK8
 
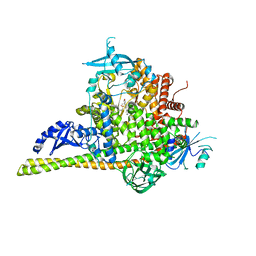 | | The co-structure of (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Mamo, M, Elling, R.A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
5SW8
 
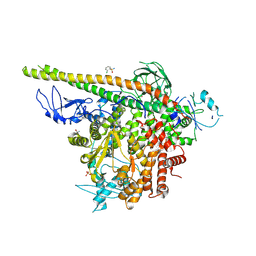 | | Crystal structure of PI3Kalpha in complex with fragments 7 and 11 | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, 2H-indazol-5-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWP
 
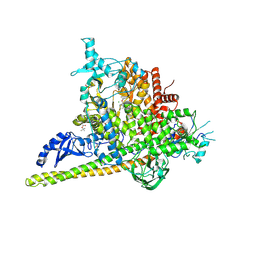 | | Crystal Structure of PI3Kalpha in complex with fragments 6 and 24 | | Descriptor: | 2-methylcyclohexane-1,3-dione, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SXB
 
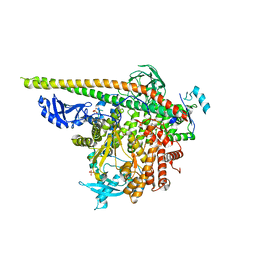 | | Crystal Structure of PI3Kalpha in complex with fragment 23 | | Descriptor: | ISATOIC ANHYDRIDE, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SXD
 
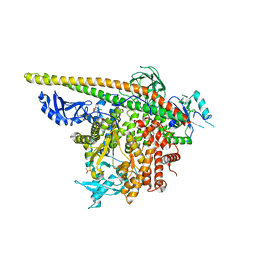 | | Crystal Structure of PI3Kalpha in complex with fragment 22 | | Descriptor: | 2-methoxybenzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWR
 
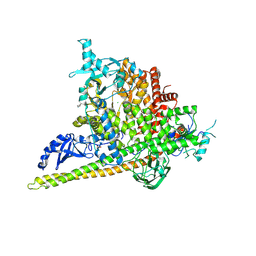 | | Crystal Structure of PI3Kalpha in complex with fragments 20 and 26 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 6-hydroxy-3,4-dihydronaphthalen-1(2H)-one, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SXA
 
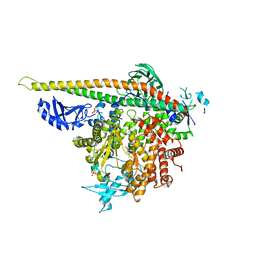 | | Crystal Structure of PI3Kalpha in complex with fragment 10 | | Descriptor: | 2-(trifluoromethyl)-1H-benzimidazol-5-amine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SXI
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 13 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, trans-cyclohexane-1,4-diol | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
2RD0
 
 | | Structure of a human p110alpha/p85alpha complex | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations.
Science, 318, 2007
|
|
2V2W
 
 | | T CELL CROSS-REACTIVITY AND CONFORMATIONAL CHANGES DURING TCR ENGAGEMENT | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, Harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, Van Der Merwe, P.A, Jones, E.Y, Mcmichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
2V2X
 
 | | T cell cross-reactivity and conformational changes during TCR engagement. | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, van der Merwe, P.A, Jones, E.Y, McMichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
2UWE
 
 | | Large CDR3a loop alteration as a function of MHC mutation | | Descriptor: | AHIII TCR ALPHA CHAIN, AHIII TCR BETA CHAIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Miller, P.J, Pazy, Y, Conti, B, Riddle, D, Biddison, W.E, Appella, E, Collins, E.J. | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
J.Mol.Biol., 373, 2007
|
|
2VYW
 
 | | Hemoglobin (Hb2) from trematode Fasciola hepatica | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dewilde, S, Ioanitescu, A.I, Kiger, L, Gilany, K, Marden, M.C, Van Doorslaer, S, Vercruysse, J, Pesce, A, Nardini, M, Bolognesi, M, Moens, L. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hemoglobins of the Trematodes Fasciola Hepatica and Paramphistomum Epiclitum: A Molecular Biological, Physico-Chemical, Kinetic, and Vaccination Study.
Protein Sci., 17, 2008
|
|
2X4N
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2VLK
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
2VLL
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | BETA-2-MICROGLOBULIN, FLU MATRIX PEPTIDE, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ishizuka, J, Stewart-Jones, G, Van Der Merwe, A, Bell, J, Mcmichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
