4UIW
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-ethyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
6IXD
 
 | | X-ray crystal structure of bPI-11 hiv-1 protease complex | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adachi, M, Hidaka, K. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Acquired Removability of Aspartic Protease Inhibitors by Direct Biotinylation.
Bioconjug.Chem., 30, 2019
|
|
4UQE
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum at 1.28 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4UIX
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 7-(3,4-dimethoxyphenyl)-N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4-oxo-4H,5H-thieno-3,2-c-pyridine-2- carboxamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 4, CALCIUM ION, ... | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UIZ
 
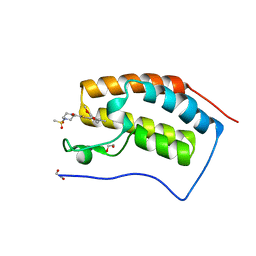 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 7-(3,4-dimethoxyphenyl)-2-(4-methanesulfonylpiperazine-1-carbonyl)-5-methyl-4H,5H-thieno-3,2-c- pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,4-dimethoxyphenyl)-5-methyl-2-(4-methylsulfonylpiperazin-1-yl)carbonyl-thieno[3,2-c]pyridin-4-one, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
7VS7
 
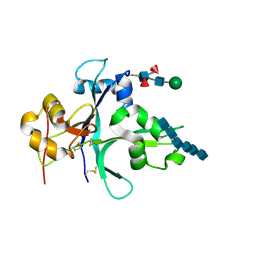 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
4UPU
 
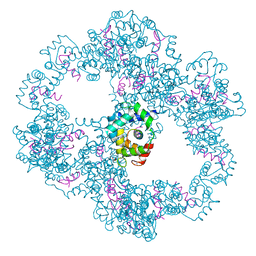 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
4UIV
 
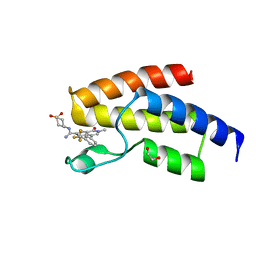 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
6JEO
 
 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
6JOU
 
 | | Crystal structure of the human nucleosome containing H2A.Z.1 S42R | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Sato, K, Mizukami, Y, Kurumizaka, H. | | Deposit date: | 2019-03-23 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-based design of an H2A.Z.1 mutant stabilizing a nucleosome in vitro and in vivo.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
6JP5
 
 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
9YFI
 
 | |
9VG0
 
 | | SIRT2 structure in complex with H3K18myr peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3.1, MYRISTIC ACID, ... | | Authors: | Zhang, N, Hao, Q. | | Deposit date: | 2025-06-12 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis of SIRT2 pre-catalysis NAD + binding dynamics and mechanism.
Rsc Chem Biol, 2025
|
|
9VGZ
 
 | |
9VG3
 
 | |
9S5G
 
 | | Temporizin-1 NMR analysis | | Descriptor: | Temporizin-1 | | Authors: | Falcigno, L, D'Auria, G, Galdiero, S, Falanga, A, Bellavita, R. | | Deposit date: | 2025-07-29 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | SOLUTION NMR | | Cite: | Temporizin-1 Meets the Membranes: Probing Membrane Inser-Tion and Disruption Mechanisms.
Antibiotics, 14, 2025
|
|
9YGK
 
 | |
9VEW
 
 | |
9U7V
 
 | | The X-ray Crystallographic Structure of oligo-1,6-glucosidase from Paenibacillus sp. STB16 | | Descriptor: | oligo-1,6-glucosidase | | Authors: | Li, Z.F, Tian, Y.X, Zhang, J.S, Ban, X.F. | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | The X-ray Crystallographic Structure of oligo-1,6-glucosidase from Paenibacillus sp. STB16
To Be Published
|
|
9OUF
 
 | |
9YFR
 
 | | perdeuterated human DJ-1, 100K | | Descriptor: | 1,2-ETHANEDIOL, Protein deglycase DJ-1 | | Authors: | Lin, J, Kovalevsky, A, Walker, A.R, Wilson, M.A. | | Deposit date: | 2025-09-26 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Environmental contributions to proton sharing in protein low barrier hydrogen bonds
To Be Published
|
|
9YCU
 
 | | Protiated human DJ-1, 100K | | Descriptor: | 1,2-ETHANEDIOL, Protein deglycase DJ-1 | | Authors: | Lin, J, Kovalevsky, A, Walker, A.R, Wilson, M.A. | | Deposit date: | 2025-09-19 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Environmental contributions to proton sharing in protein low barrier hydrogen bonds
To Be Published
|
|
9RC4
 
 | |
9RNT
 
 | |
