7PMQ
 
 | |
1YNE
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | APOLIPOPROTEIN B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YNG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1YLG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1MHK
 
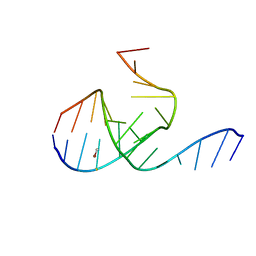 | | Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn | | Descriptor: | BROMIDE ION, RNA 12-mer BCh12, RNA 14-mer BCh12 | | Authors: | Szep, S, Wang, J, Moore, P.B. | | Deposit date: | 2002-08-20 | | Release date: | 2002-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a 26-nucleotide RNA containing a hook-turn
RNA, 9, 2003
|
|
7PLI
 
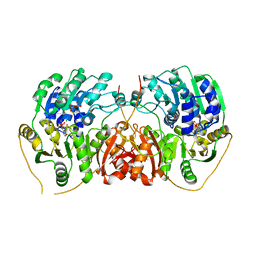 | | DEAD-box helicase DbpA bound to single stranded RNA and ADP/BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DbpA, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wurm, J.P. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA-duplex unwinding by the DEAD-box helicase DbpA.
Rna, 29, 2023
|
|
6GKH
 
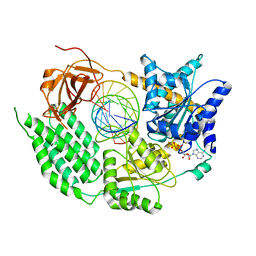 | | CryoEM structure of the MDA5-dsRNA filament in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-05-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6GKM
 
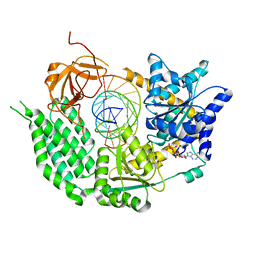 | | CryoEM structure of the MDA5-dsRNA filament in complex with ATP (10 mM) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-05-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6H66
 
 | | CryoEM structure of the MDA5-dsRNA filament with 93 degree twist and without nucleotide | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6EZD
 
 | |
3HL2
 
 | | The crystal structure of the human SepSecS-tRNASec complex | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Monothiophosphate, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Palioura, S, Steitz, T.A, Soll, D, Simonovic, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation.
Science, 325, 2009
|
|
6GJZ
 
 | | CryoEM structure of the MDA5-dsRNA filament in complex with AMPPNP | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6H61
 
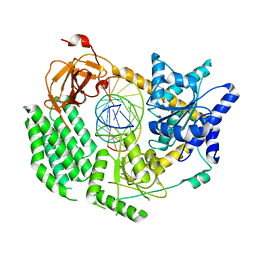 | | CryoEM structure of the MDA5-dsRNA filament with 89 degree twist and without nucleotide | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-07-25 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
4XBF
 
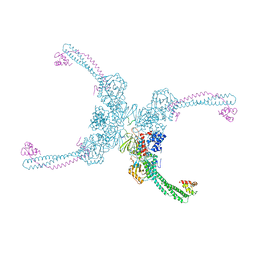 | | Structure of LSD1:CoREST in complex with ssRNA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Luka, Z, Loukachevitch, L.V, Martin, W.J, Wagner, C, Reiter, N.J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | G-quadruplex RNA binding and recognition by the lysine-specific histone demethylase-1 enzyme.
RNA, 22, 2016
|
|
6G1X
 
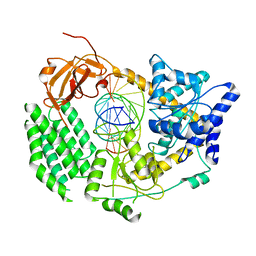 | | CryoEM structure of the MDA5-dsRNA filament with 91-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6G1S
 
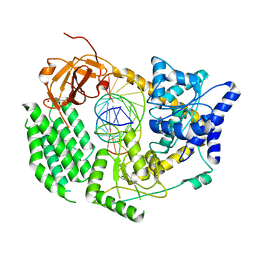 | | CryoEM structure of the MDA5-dsRNA filament with 87-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6G19
 
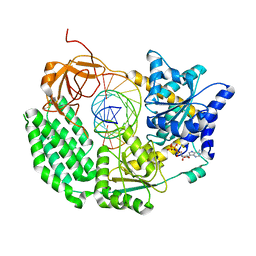 | | CryoEM structure of the MDA5-dsRNA filament with 74-degree helical twist | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*CP*AP*AP*GP*CP*CP*GP*AP*GP*GP*AP*GP*AP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
5Z5E
 
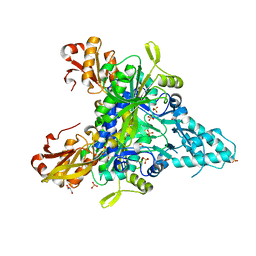 | |
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | Descriptor: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | Authors: | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
4TS1
 
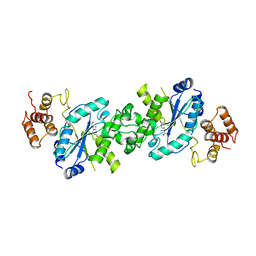 | |
2JAN
 
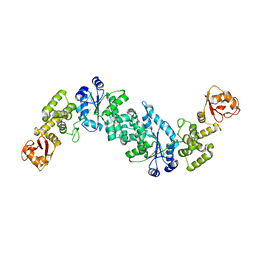 | |
7YFY
 
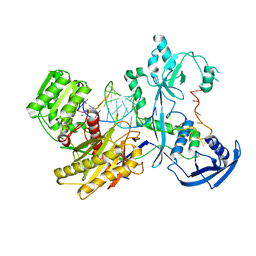 | | Cryo-EM structure of the Mili-piRNA- target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, RNA (5'-R(P*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-09 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
6AGT
 
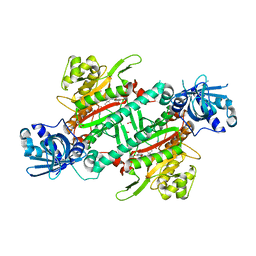 | | Crystal structure of PfKRS complexed with chromone inhibitor | | Descriptor: | COBALT (II) ION, FORMIC ACID, LYSINE, ... | | Authors: | Yogavel, M, Sharma, A, Sharma, A, Baragana, B, Walpole, C. | | Deposit date: | 2018-08-14 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
9JVN
 
 | | Z-FORM DNA-RNA HYBRID | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*CP*GP*CP*G)-3'), RNA (5'-R(*CP*GP*CP*GP*UP*GP*CP*G)-3') | | Authors: | Wang, S, Xu, Y. | | Deposit date: | 2024-10-09 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | SOLUTION NMR | | Cite: | Z-form DNA-RNA hybrid blocks DNA replication.
Nucleic Acids Res., 53, 2025
|
|
6J7Z
 
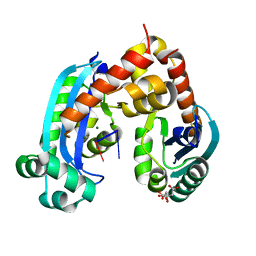 | | Human mitochondrial Oligoribonuclease in complex with RNA | | Descriptor: | CITRIC ACID, MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Chu, L.Y, Agrawal, S, Yuan, H.S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural insights into nanoRNA degradation by human Rexo2.
Rna, 25, 2019
|
|
