3QRY
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 1-deoxymannojirimycin complex | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-18 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QEX
 
 | |
3QNO
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite 3tCo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Polymerase, ... | | Authors: | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-02-08 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
3QX9
 
 | | Crystal structure of MID domain from hAGO2 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein argonaute-2 | | Authors: | Frank, F, Fabian, M.R, Stepinski, J, Jemielity, J, Darzynkiewicz, E, Sonenberg, N, Nagar, B. | | Deposit date: | 2011-03-01 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of 5'-mRNA-cap interactions with the human AGO2 MID domain.
Embo Rep., 12, 2011
|
|
3Q80
 
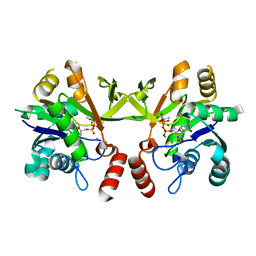 | | Structure of Mtb 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (IspD) Complexed with CDP-ME | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, CHLORIDE ION, ... | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Ioerger, T.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-01-05 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis 2-C-methyl-D-erythritol 4-phosphate cytidyltransferase (IspD): a candidate antitubercular drug target
Proteins, 2011
|
|
3QPF
 
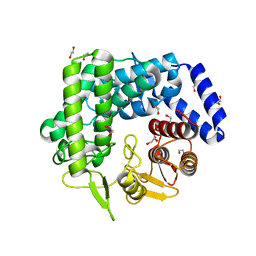 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Streptococcus pneumoniae SP_2144 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J. | | Deposit date: | 2011-02-12 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3Q5K
 
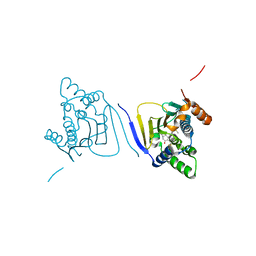 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
3Q9H
 
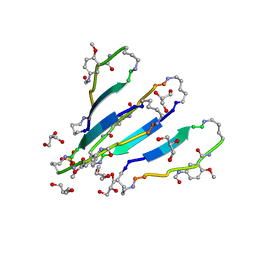 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | 1,4-BUTANEDIOL, Cyclic pseudo-peptide LVFFA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3QEI
 
 | | RB69 DNA Polymerase (L561A/S565G/Y567A) Ternary Complex with dCTP Opposite Difluorotoluene Nucleoside | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-01-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.178 Å) | | Cite: | Structural Basis for Differential Insertion Kinetics of dNMPs Opposite a Difluorotoluene Nucleotide Residue.
Biochemistry, 51, 2012
|
|
1TF8
 
 | | Streptomyces griseus aminopeptidase complexed with L-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of inhibitory aromatic amino acids to Streptomyces griseus aminopeptidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T16
 
 | | Crystal structure of the bacterial fatty acid transporter FadL from Escherichia coli | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, COPPER (II) ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | van den Berg, B, Black, P.N, Clemons Jr, W.M, Rapoport, T.A. | | Deposit date: | 2004-04-15 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the long-chain fatty acid transporter FadL.
Science, 304, 2004
|
|
1T7P
 
 | | T7 DNA POLYMERASE COMPLEXED TO DNA PRIMER/TEMPLATE,A NUCLEOSIDE TRIPHOSPHATE, AND ITS PROCESSIVITY FACTOR THIOREDOXIN | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*CP*TP*TP*GP*GP*CP*AP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*2DA)-3'), ... | | Authors: | Doublie, S, Tabor, S, Long, A.M, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1997-09-24 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution.
Nature, 391, 1998
|
|
1TKF
 
 | | Streptomyces griseus aminopeptidase complexed with D-tryptophan | | Descriptor: | Aminopeptidase, CALCIUM ION, D-TRYPTOPHAN, ... | | Authors: | Reiland, V, Gilboa, R, Spungin-Bialik, A, Schomburg, D, Shoham, Y, Blumberg, S, Shoham, G. | | Deposit date: | 2004-06-08 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactions of D Amino Acids with Streptomyces griseus Aminopeptidase
To be Published
|
|
1TLZ
 
 | | Tsx structure complexed with uridine | | Descriptor: | Nucleoside-specific channel-forming protein tsx, URIDINE | | Authors: | Ye, J, van den Berg, B. | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the bacterial nucleoside transporter Tsx.
Embo J., 23, 2004
|
|
1VS0
 
 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
1VQJ
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 (V35I) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VRZ
 
 | | Helix turn helix motif | | Descriptor: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | Authors: | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
8FFT
 
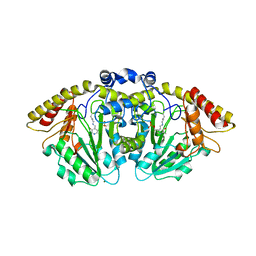 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine | | Descriptor: | Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Lima, S.T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8FFU
 
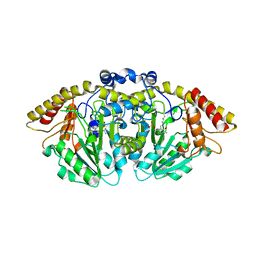 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine, with the substrate bound | | Descriptor: | (2S,4S)-5-carbamimidamido-4-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pentanoic acid (non-preferred name), Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
6RSQ
 
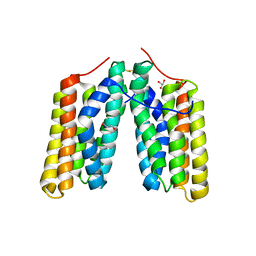 | | Helical folded domain of mouse CAP1 | | Descriptor: | Adenylyl cyclase-associated protein 1, GLYCEROL | | Authors: | Kotila, T, Kogan, K, Lappalainen, P. | | Deposit date: | 2019-05-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin.
Nat Commun, 10, 2019
|
|
6RUM
 
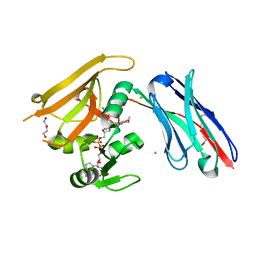 | | Crystal structure of GFP-LAMA-G97 - a GFP enhancer nanobody with cpDHFR insertion and TMP and NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GFP-LAMA-G97 a GFP enhancer nanobody with cpDHFR insertion, ... | | Authors: | Farrants, H, Tarnawski, M, Mueller, T.G, Otsuka, S, Hiblot, J, Koch, B, Kueblbeck, M, Kraeusslich, H.-G, Ellenberg, J, Johnsson, K. | | Deposit date: | 2019-05-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemogenetic Control of Nanobodies.
Nat.Methods, 17, 2020
|
|
6S23
 
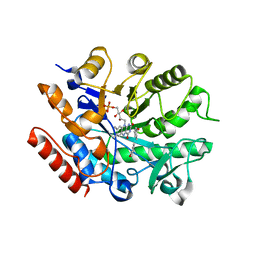 | | Crystal structure of ene-reductase GsOYE from Galleria sulphuraria in complex with 2-methyl-cyclopenten-1-one | | Descriptor: | 2-methylcyclopenta-2,4-dien-1-one, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Robescu, M.S, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6S4C
 
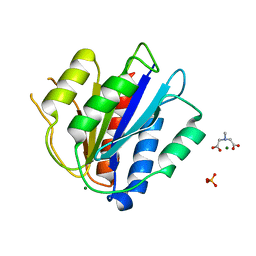 | | Crystal Structure of the vWFA2 subdomain of type VII collagen | | Descriptor: | Collagen alpha-1(VII) chain, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gebauer, J.M, Flachsenberg, F, Baumann, U, Seeger, K. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterization of the type VII collagen vWFA2 subdomain leads to identification of two binding sites.
Febs Open Bio, 10, 2020
|
|
6S31
 
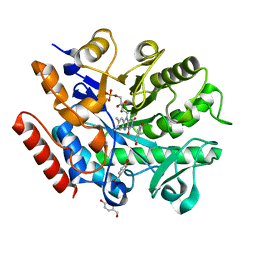 | | Crystal structure of ene-reductase GsOYE from Galdieria sulphuraria in complex with 4-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH2 dehydrogenase-like protein, P-HYDROXYBENZALDEHYDE | | Authors: | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6S8Y
 
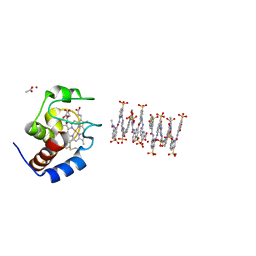 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
