4CA9
 
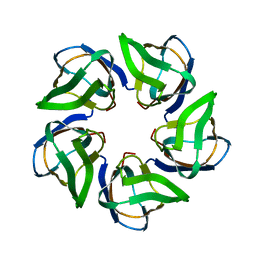 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
8IF5
 
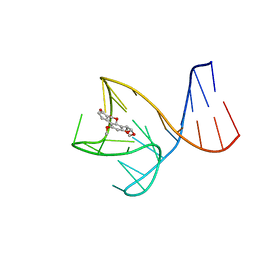 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
3F2E
 
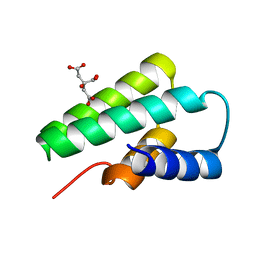 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
5HAW
 
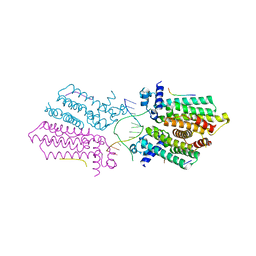 | |
5HBY
 
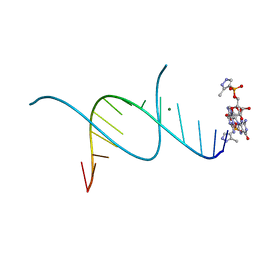 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue-3 binding sites | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*C)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Wang, J, Szostak, J.W. | | Deposit date: | 2016-01-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
6A28
 
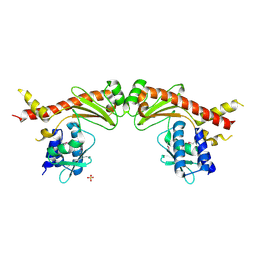 | | Crystal structure of PprA W183R mutant form 2 | | Descriptor: | DNA repair protein PprA, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
6EC0
 
 | | Crystal structure of the wild-type heterocomplex between coil 1B domains of human intermediate filament proteins keratin 1 (KRT1) and keratin 10 (KRT10) | | Descriptor: | CADMIUM ION, COBALT (II) ION, Keratin 1, ... | | Authors: | Eldirany, S.A, Lomakin, I.B, Bunick, C.G. | | Deposit date: | 2018-08-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Human keratin 1/10-1B tetramer structures reveal a knob-pocket mechanism in intermediate filament assembly.
Embo J., 38, 2019
|
|
4OI0
 
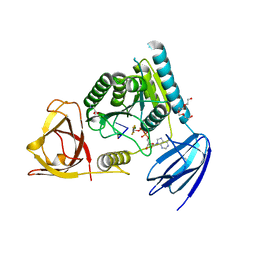 | | bound to ssRNA dinucleotide GC, ADP, AlF4-, and Mg2+(transition state, data set I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
8QJG
 
 | | Crystal structure of cytochrome domain 2 from PgcA | | Descriptor: | 1,2-ETHANEDIOL, HEME C, Lipoprotein cytochrome c, ... | | Authors: | Nash, B.W, Edwards, M.J, Clarke, T.A. | | Deposit date: | 2023-09-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tethered heme domains in a triheme cytochrome allow for increased electron transport distances.
Protein Sci., 33, 2024
|
|
8EKY
 
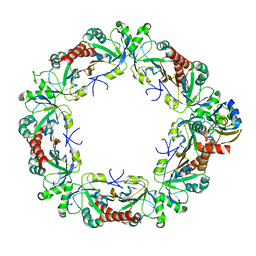 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
6NKX
 
 | |
1NZ5
 
 | | The Horse heart myoglobin variant K45E/K63E complexed with Manganese | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5MGX
 
 | |
8EKW
 
 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
4GKL
 
 | |
6B2X
 
 | | Apo YiuA Crystal Form 1 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute-binding periplasmic protein of iron/siderophore ABC transporter | | Authors: | Radka, C.D, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2017-09-20 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of the Yersinia pestis iron chaperone YiuA reveals a basic triad binding motif for the chelated metal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7T5H
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
6NKR
 
 | | Ternary complex crystal structure of K289M variant of DNA polymerase Beta with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Revealing an Internal Stabilization Deficiency in the DNA Polymerase beta K289M Cancer Variant through the Combined Use of Chemical Biology and X-ray Crystallography.
Biochemistry, 59, 2020
|
|
6NKU
 
 | |
6BVV
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
4YRW
 
 | | rat xanthine oxidoreductase, C-terminal deletion protein variant | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
1OAG
 
 | |
1BI4
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-22 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
5IVL
 
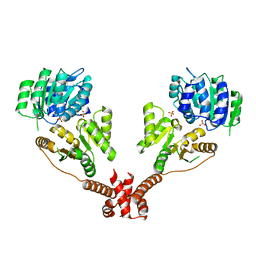 | | CshA Helicase | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA, SULFATE ION | | Authors: | Huen, J, Lin, C.-L, Yi, W.-L, Li, C.-L, Yuan, H. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into a Unique Dimeric DEAD-Box Helicase CshA that Promotes RNA Decay.
Structure, 25, 2017
|
|
5IW3
 
 | | anti-CD20 monoclonal antibody Fc fragment | | Descriptor: | ACETATE ION, Ig gamma-1 chain C region, SULFATE ION, ... | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of anti-CD20 monoclonal antibody Fc fragment at 2.05 Angstroms resolution
To Be Published
|
|
