4V65
 
 | | Structure of the E. coli ribosome in the Pre-accommodation state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Devkota, B, Caulfield, T.R, Tan, R.-Z, Harvey, S.C. | | Deposit date: | 2008-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Structure of the E. coli Ribosome Before and After Accommodation: Implications for Proofreading
To be Published
|
|
2DTZ
 
 | |
7LWE
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-7629 | | Descriptor: | ACETATE ION, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWF
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-9320 | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, N-(3-chloropyridin-4-yl)-2-[5-(3-cyano-4-hydroxyphenyl)-3-methyl-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWG
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12694 | | Descriptor: | 5-{(5S)-1-[2-({3-chloro-6-[(2S)-2,4-dimethylpiperazin-1-yl]-2-fluoropyridin-4-yl}amino)-2-oxoethyl]-4-oxo-4,6,7,8-tetrahydro-1H-dipyrrolo[1,2-a:2',3'-d]pyrimidin-3-yl}-3,4-difluoro-2-hydroxybenzamide, B-cell lymphoma 6 protein, GLYCEROL, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7ZVI
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression | | Descriptor: | Orf22, Sri | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Marina, A, Penades, J.R. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.973 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
7EPG
 
 | |
7EPI
 
 | |
7EPJ
 
 | |
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
7PWJ
 
 | | dUTPase from human in complex with Stl | | Descriptor: | 1,2-ETHANEDIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | Authors: | Sanz-Frasquet, C, Marina, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
6F5M
 
 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4P
 
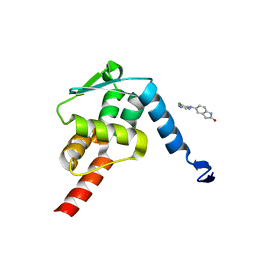 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
8SSU
 
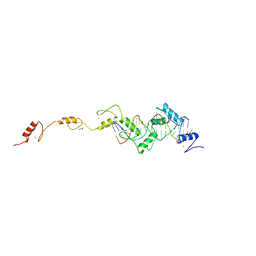 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SST
 
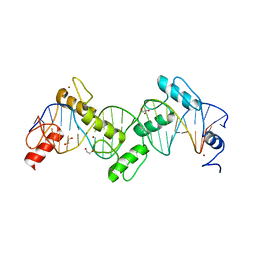 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
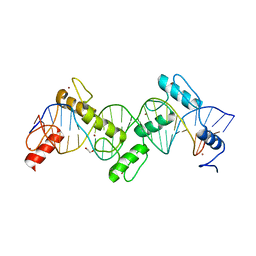 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | Descriptor: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | Descriptor: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SVD
 
 | |
7RTE
 
 | | X-ray structure of wild type RBPJ-L3MBTL3-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Hall, D.P, Kovall, R.A, Yuan, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.
Nucleic Acids Res., 50, 2022
|
|
8C7S
 
 | | Transcriptional pleiotropic repressor CodY from Staphylococcus aureus in complex with Ile, GTP, and a 30-bp DNA fragment encompassing two overlapping binding sites | | Descriptor: | DNA (30-MER), GUANOSINE-5'-TRIPHOSPHATE, Global transcriptional regulator CodY (Fragment), ... | | Authors: | Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2023-01-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into CodY activation and DNA recognition.
Nucleic Acids Res., 51, 2023
|
|
