2CCR
 
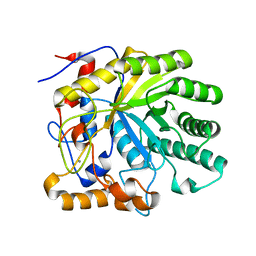 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, TRIETHYLENE GLYCOL, YVFO, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
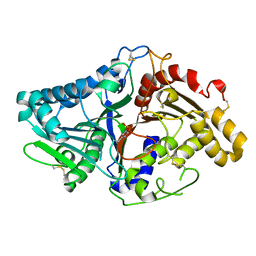
1V0V
 
 | |
4GT0
 
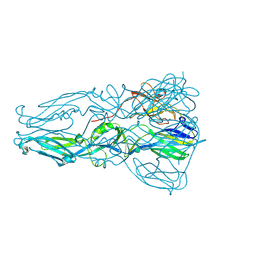 | | Structure of dengue virus serotype 1 sE containing stem to residue 421 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Klein, D.E, Choi, J.L, Harrison, S.C. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of a dengue virus envelope protein late-stage fusion intermediate.
J.Virol., 87, 2013
|
|
3SEW
 
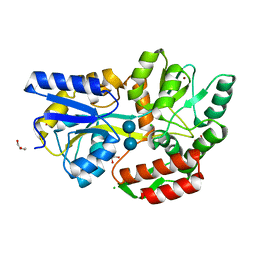 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form I) | | Descriptor: | CHLORIDE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
2PIK
 
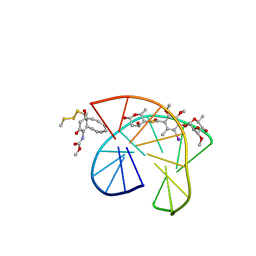 | | CALICHEAMICIN GAMMA1I-DNA COMPLEX, NMR, 6 STRUCTURES | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-31 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calicheamicin gamma 1I-DNA complex.
J.Mol.Biol., 265, 1997
|
|
1NQG
 
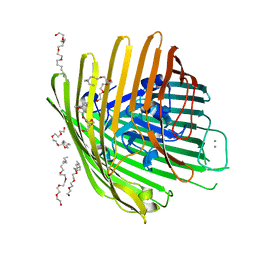 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI, WITH BOUND CALCIUM | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, vitamin b12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
1AOL
 
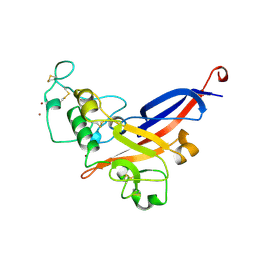 | | FRIEND MURINE LEUKEMIA VIRUS RECEPTOR-BINDING DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GP70, ZINC ION | | Authors: | Fass, D, Davey, R.A, Hamson, C.A, Kim, P.S, Cunningham, J.M, Berger, J.M. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution.
Science, 277, 1997
|
|
2A2U
 
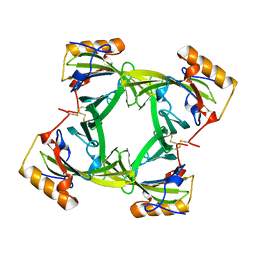 | |
2A3E
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, SULFATE ION, ... | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
2AFY
 
 | | Formylglycine generating enzyme C341S mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Sulfatase modifying factor 1 | | Authors: | Roeser, D, Rudolph, M.G. | | Deposit date: | 2005-07-26 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1RJP
 
 | | Crystal structure of D-aminoacylase in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
3SEY
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SER
 
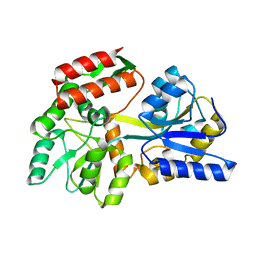 | | Zn-mediated Polymer of Maltose-binding Protein K26H/K30H by Synthetic Symmetrization | | Descriptor: | CALCIUM ION, CHLORIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
2A42
 
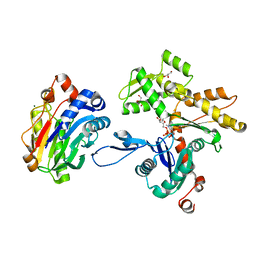 | | Actin-DNAse I Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-06-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1R1O
 
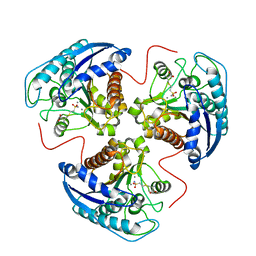 | |
1R4D
 
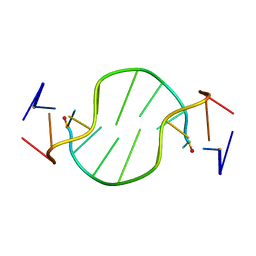 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
4JZZ
 
 | |
3NQS
 
 | | Crystal Structure of Inducible Nitric Oxide Synthase with N-Nitrosated-pterin | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, ... | | Authors: | Rosenfeld, R.J, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2010-06-29 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitric-oxide synthase forms N-NO-pterin and S-NO-cys: implications for activity, allostery, and regulation.
J.Biol.Chem., 285, 2010
|
|
1RII
 
 | | Crystal structure of phosphoglycerate mutase from M. Tuberculosis | | Descriptor: | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, GLYCEROL | | Authors: | Mueller, P, Sawaya, M.R, Chan, S, Wu, Y, Pashkova, I, Perry, J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.70 angstroms X-ray crystal structure of Mycobacterium tuberculosis phosphoglycerate mutase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SA9
 
 | | Crystal Structure of the RNA octamer GGCGAGCC | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*GP*CP*C)-3' | | Authors: | Jang, S.B, Baeyens, K, Jeong, M.S, SantaLucia Jr, J, Turner, D, Holbrook, S.R. | | Deposit date: | 2004-02-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structures of two RNA octamers containing tandem G.A base pairs.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ICG
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[F]U IN PRESENCE OF IR(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UFP))-3', CHLORIDE ION, IRIDIUM HEXAMMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-03-31 | | Release date: | 2001-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4JZW
 
 | |
1RJR
 
 | | The crystal structure of the D-aminoacylase D366A mutant in complex with 100mM ZnCl2 | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1KTB
 
 | | The Structure of alpha-N-Acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Garman, S.C, Hannick, L, Zhu, A, Garboczi, D.N. | | Deposit date: | 2002-01-15 | | Release date: | 2002-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of alpha-N-acetylgalactosaminidase: molecular basis of glycosidase deficiency diseases
Structure, 10, 2002
|
|
4H44
 
 | | 2.70 A Cytochrome b6f Complex Structure From Nostoc PCC 7120 | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Hasan, S.S, Yamashita, E, Baniulis, D, Cramer, W.A. | | Deposit date: | 2012-09-16 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
