6HY8
 
 | |
6O2Z
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 32 | | Descriptor: | 6-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methylpyridine-3-carbonitrile, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6NOH
 
 | | Crystal structure of FBF-2 repeat 5 mutant (C363S, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NOD
 
 | | Crystal structure of C. elegans PUF-8 in complex with RNA | | Descriptor: | PUF (Pumilio/FBF) domain-containing, RNA (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3') | | Authors: | Wang, Y, McCann, K.L, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NT2
 
 | | type 1 PRMT in complex with the inhibitor GSK3368715 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, N~1~-({5-[4,4-bis(ethoxymethyl)cyclohexyl]-1H-pyrazol-4-yl}methyl)-N~1~,N~2~-dimethylethane-1,2-diamine, ... | | Authors: | Concha, N.O. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss.
Cancer Cell, 36, 2019
|
|
6HNM
 
 | | Crystal structure of IdmH 96-104 loop truncation variant | | Descriptor: | putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6NRP
 
 | | Putative short-chain dehydrogenase/reductase (SDR) from Acinetobacter baumannii | | Descriptor: | 3-oxoacyl-ACP reductase FabG | | Authors: | Cross, E.M, Smith, K.M, Shaw, K.I, Aragao, D, Forwood, J.K. | | Deposit date: | 2019-01-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Acinetobacter baumannii fatty acid synthesis 3-oxoacyl-ACP reductases.
Sci Rep, 11, 2021
|
|
6NVO
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
6NOF
 
 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NVQ
 
 | |
8W4Q
 
 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4R
 
 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
6N0P
 
 | |
6N0K
 
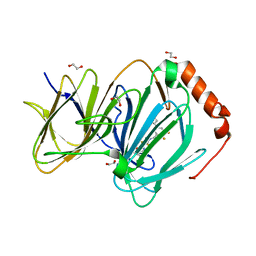 | | The complex of CCG-257081 bound to pirin | | Descriptor: | (3R)-N-(4-chlorophenyl)-5,5-difluoro-1-[3-fluoro-5-(pyridin-4-yl)benzene-1-carbonyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Lisabeth, E.M, Jin, X, Neubig, R. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Antifibrotic and Antimetastatic Compounds
ACS Pharmacol Transl Sci, 2, 2019
|
|
6HNN
 
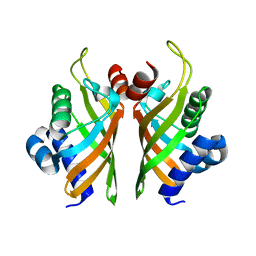 | | Crystal structure of wild-type IdmH, a putative polyketide cyclase from Streptomyces antibioticus | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
6I89
 
 | |
6NOC
 
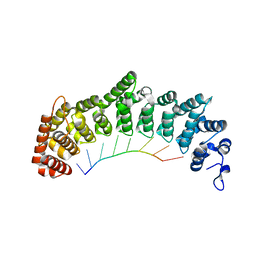 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y) in complex with 8-nt RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6H4I
 
 | | Usp28 catalytic domain apo | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4H
 
 | | Usp28 catalytic domain variant E593D in complex with UbPA | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28, ... | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4K
 
 | | Structure of the Usp25 C-terminal domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6NVP
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
6NYH
 
 | | Structure of human RIPK1 kinase domain in complex with GNE684 | | Descriptor: | (5S)-N-[(3S)-7-methoxy-1-methyl-2-oxo-2,3,4,5-tetrahydro-1H-pyrido[3,4-b]azepin-3-yl]-5-phenyl-6,7-dihydro-5H-pyrrolo[1,2-b][1,2,4]triazole-2-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases.
Cell Death Differ., 27, 2020
|
|
6O2Y
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 24 | | Descriptor: | 4-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methoxybenzonitrile, BETA-MERCAPTOETHANOL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6HY4
 
 | |
8ZMU
 
 | |
