6WKQ
 
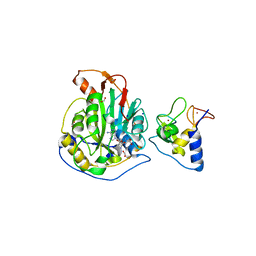 | | 1.98 Angstrom Resolution Crystal Structure of NSP16-NSP10 Heterodimer from SARS-CoV-2 in Complex with Sinefungin | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
8HFH
 
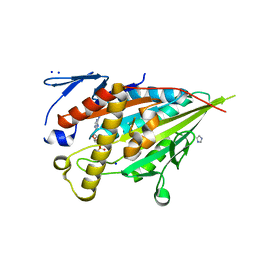 | | CENP-E motor domain in complex with AMPPNP and Mg2+ | | Descriptor: | Centromere-associated protein E, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Shibuya, A, Yokoyama, H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the motor domain of centromere-associated protein E in complex with a non-hydrolysable ATP analogue.
Febs Lett., 597, 2023
|
|
3WXU
 
 | |
7EXX
 
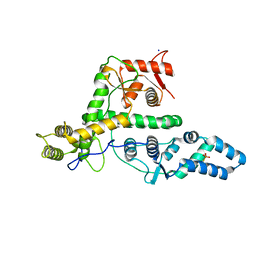 | | The structure of DndG | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptG, SODIUM ION | | Authors: | Wu, D, Wang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
3OQ2
 
 | | Structure of a CRISPR associated protein Cas2 from Desulfovibrio vulgaris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Samai, P, Smith, P, Shuman, S. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7EZW
 
 | | Cyclic Peptide that Interacts with the eIF4E Capped-mRNA Binding Site | | Descriptor: | ALA-CYS-GLU-MET-GLY-PHE-PHE-GLN-ASP-CYS-GLY, Eukaryotic translation initiation factor 4E, SODIUM ION | | Authors: | Brown, C.J, Ng, S, Frosi, Y. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
7F0N
 
 | |
7F4X
 
 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
8HAP
 
 | | Crystal structure of thermostable acetaldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, SODIUM ION, ... | | Authors: | Mine, S, Nakabayashi, M, Ishikawa, K. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable acetaldehyde dehydrogenase from the hyperthermophilic archaeon Sulfolobus tokodaii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3OT1
 
 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
3PBJ
 
 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | Descriptor: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | Authors: | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-10-20 | | Release date: | 2011-11-30 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
3PDV
 
 | |
8H9O
 
 | |
3PC4
 
 | |
8H9X
 
 | |
8HA1
 
 | |
7FCR
 
 | |
7FCS
 
 | |
3PBX
 
 | |
7FDN
 
 | |
8H1G
 
 | |
3PK8
 
 | | Urate oxidase under 0.5 MPa / 5 bars pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
