6OWW
 
 | | Crystal structure of a Human Cardiac Calsequestrin Filament Complexed with Ytterbium | | Descriptor: | Calsequestrin-2, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8H58
 
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
6LSS
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state preA | | Descriptor: | 28S rRNA, 5S rRNA, 60S ribosomal protein L11, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6Y1X
 
 | | X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Sosa-Fajardo, A, Legrand, P, Paya-Tormo, L, Martin, L, Pellicer-Martinez, M.T, Echavarri-Erasun, C, Vernede, X, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Mechanism of the Radical SAM Carbide Synthase NifB, a Key Nitrogenase Cofactor Maturating Enzyme.
J.Am.Chem.Soc., 142, 2020
|
|
8CQH
 
 | |
8PKL
 
 | |
8PEG
 
 | |
7POM
 
 | |
4XI2
 
 | |
6PIB
 
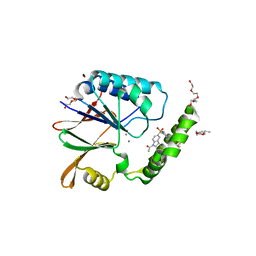 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SGK
 
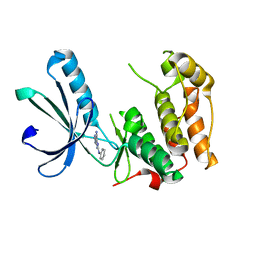 | | Nek2 kinase bound to inhibitor 102 | | Descriptor: | 2-phenylazanyl-9~{H}-purine-6-carbonitrile, Serine/threonine-protein kinase Nek2 | | Authors: | Richards, M.W, Mas-Droux, C.P, Bayliss, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Arylamino-6-ethynylpurines are cysteine-targeting irreversible inhibitors of Nek2 kinase.
Rsc Med Chem, 11, 2020
|
|
6ANZ
 
 | |
4L1T
 
 | |
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | Descriptor: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
8UO0
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class P) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | Descriptor: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
8UNN
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class C) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNL
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class A) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNS
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class H) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNR
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class G) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNX
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class M) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UNW
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class L) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UO2
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class R) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8UO1
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class Q) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
