8BQX
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BTR
 
 | | Giardia Ribosome in PRE-T Hybrid State (D2) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BQD
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BR8
 
 | | Giardia ribosome in POST-T state (A1) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BRM
 
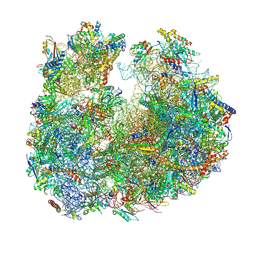 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BUU
 
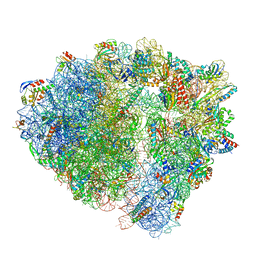 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-11-30 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
8BHL
 
 | |
8BHP
 
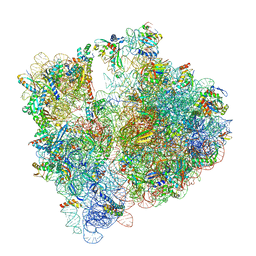 | |
8BHF
 
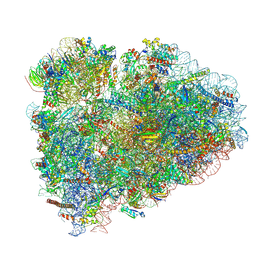 | | Cryo-EM structure of stalled rabbit 80S ribosomes in complex with human CCR4-NOT and CNOT4 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S11, ... | | Authors: | Absmeier, E, Chandrasekaran, V, O'Reilly, F.J, Stowell, J.A.W, Rappsilber, J, Passmore, L.A. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHN
 
 | |
8BH4
 
 | |
8BGE
 
 | |
7UVY
 
 | | A. baumannii ribosome-Streptothricin-D complex: 70S with P-site tRNA | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
7UVW
 
 | | A. baumannii ribosome: 70S with E-site tRNA | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYJ
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
