4HTP
 
 | |
2D4U
 
 | | Crystal Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr | | Descriptor: | Methyl-accepting chemotaxis protein I | | Authors: | Imada, K, Tajima, H, Namba, K, Sakuma, M, Homma, M, Kawagishi, I. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in the bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 2011
|
|
1I8E
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND A22 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND A22 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-13 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1I98
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D18 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D18 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-18 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
2YN6
 
 | | Pentameric Ligand-Gated Ion Channel ELIC in Complex with Barium | | Descriptor: | BARIUM ION, PENTAMERIC LIGAND-GATED ION CHANNEL ELIC | | Authors: | Zimmermann, I, Marabelli, A, Bertozzi, C, Sivilotti, L.G, Dutzler, R. | | Deposit date: | 2012-10-12 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Inhibition of the Prokaryotic Pentameric Ligand-Gated Ion Channel Elic by Divalent Cations.
Plos Biol., 10, 2012
|
|
2YOE
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with GABA and flurazepam | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, Flurazepam, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2012-10-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1I6Y
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND A1 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND A1 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-06 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
4OTG
 
 | | Crystal Structure of PRK1 Catalytic Domain in Complex with Lestaurtinib | | Descriptor: | Lestaurtinib, Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
4OTH
 
 | | Crystal Structure of PRK1 Catalytic Domain in Complex with Ro-31-8220 | | Descriptor: | BISINDOLYLMALEIMIDE IX, Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
4OTD
 
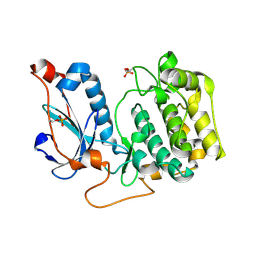 | | Crystal Structure of PRK1 Catalytic Domain | | Descriptor: | Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
1I93
 
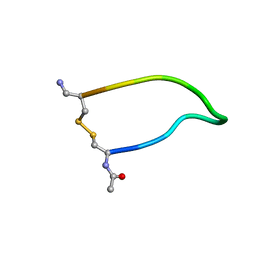 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D16 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D16 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1VQZ
 
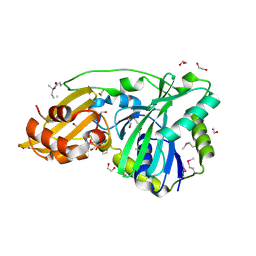 | |
1VS0
 
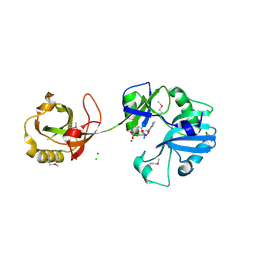 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
4DZH
 
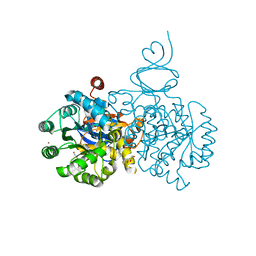 | | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn | | Descriptor: | AMIDOHYDROLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Raushel, F.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Crystal structure of an adenosine deaminase from xanthomonas campestris (target nysgrc-200456) with bound zn
to be published
|
|
5DKA
 
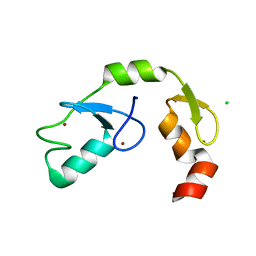 | | A C2HC zinc finger is essential for the activity of the RING ubiquitin ligase RNF125 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF125, MAGNESIUM ION, ... | | Authors: | Boer, D.R, Coll, M, Bijlmakers, M.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125.
Sci Rep, 6, 2016
|
|
7EV3
 
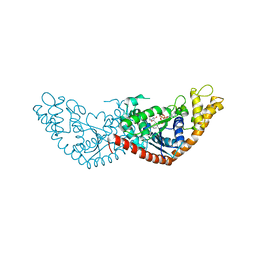 | |
7EV2
 
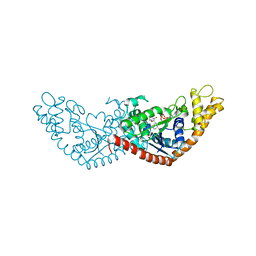 | |
6AGY
 
 | | Aspergillus fumigatus Af293 NDK | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Hu, Y, Han, L. | | Deposit date: | 2018-08-15 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of crystal structure and key residues of Aspergillus fumigatus nucleoside diphosphate kinase.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
2C8M
 
 | |
7P47
 
 | | Structure of the E3 ligase Smc5/Nse2 in complex with Ubc9-SUMO thioester mimetic | | Descriptor: | E3 SUMO-protein ligase MMS21, SUMO-conjugating enzyme UBC9, Structural maintenance of chromosomes protein 5, ... | | Authors: | Lascorz, J, Varejao, N, Reverter, D. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Nat Commun, 12, 2021
|
|
6FGA
 
 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
1IK9
 
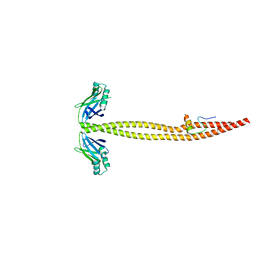 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
2CMN
 
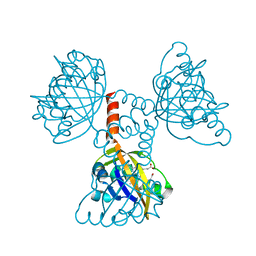 | | A Proximal Arginine Residue in the Switching Mechanism of the FixL Oxygen Sensor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gilles-Gonzalez, M.-A, Caceres, A.I, Silva Sousa, E.H, Tomchick, D.R, Brautigam, C.A, Gonzalez, C, Machius, M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Proximal Arginine R206 Participates in Switching of the Bradyrhizobium Japonicum Fixl Oxygen Sensor
J.Mol.Biol., 360, 2006
|
|
2ARU
 
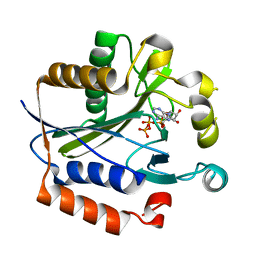 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2CGH
 
 | |
