1UGK
 
 | | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342) | | Descriptor: | Synaptotagmin IV | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342)
To be Published
|
|
1DB1
 
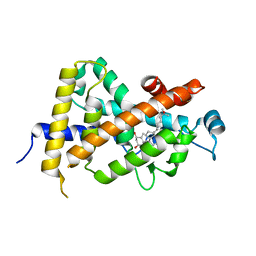 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | Authors: | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
5Y30
 
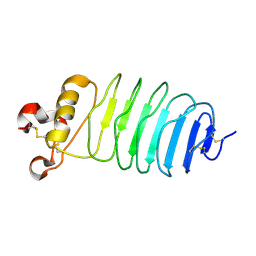 | | Crystal structure of LGI1 LRR domain | | Descriptor: | Leucine-rich glioma-inactivated protein 1 | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
4UFW
 
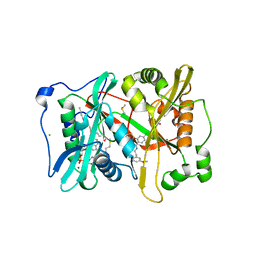 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 22) | | Descriptor: | 2-oxopentadecyl-CoA, 4-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, CHLORIDE ION, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
5V57
 
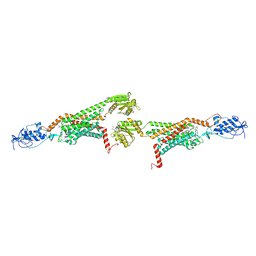 | | 3.0A SYN structure of the multi-domain human smoothened receptor in complex with TC114 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
6FI0
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6ISB
 
 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4UFX
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 19) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
1UFM
 
 | | Solution structure of the PCI domain | | Descriptor: | COP9 complex subunit 4 | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Shirouzu, M, Hayashizaki, Y, The RIKEN Genome Exploration Research Group Phase I & II Teams, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PCI domain
To be Published
|
|
1UGV
 
 | | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621) | | Descriptor: | Olygophrenin-1 like protein | | Authors: | Inoue, K, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621)
To be Published
|
|
3HT8
 
 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1UF0
 
 | | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase | | Descriptor: | Serine/threonine-protein kinase DCAMKL1 | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal DCX domain of human doublecortin-like kinase
To be Published
|
|
1UFZ
 
 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
3HU8
 
 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
4UFV
 
 | | Plasmodium vivax N-myristoyltransferase in complex with a pyridyl inhibitor (compound 18) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yu, Z, Brannigan, J.A, Rangachari, K, Heal, W.P, Wilkinson, A.J, Holder, A.A, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-03-19 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Pyridyl-Based Inhibitors of Plasmodium Falciparum N-Myristoyltransferase
Medchemcomm, 6, 2015
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
1UDY
 
 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
5V56
 
 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
1UEW
 
 | | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) | | Descriptor: | MEMBRANE ASSOCIATED GUANYLATE KINASE INVERTED-2 (MAGI-2) | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein)
To be Published
|
|
5Y2Z
 
 | | Crystal structure of human LGI1 EPTP-ADAM22 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
1UG7
 
 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | Descriptor: | 2610208M17Rik protein | | Authors: | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-13 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
6U30
 
 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
3HU9
 
 | | Nitrosobenzene in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, NITROSOBENZENE, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
