2EUM
 
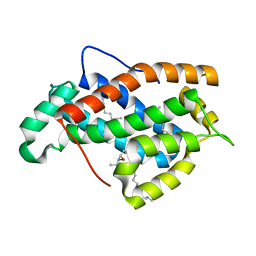 | | Crystal structure of human Glycolipid Transfer Protein complexed with 8:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
1GUO
 
 | |
2JGS
 
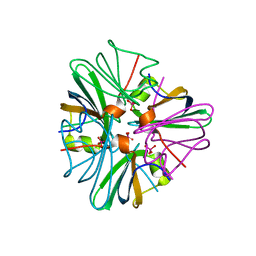
 | | Circular permutant of avidin | | Descriptor: | BIOTIN, CIRCULAR PERMUTANT OF AVIDIN | | Authors: | Maatta, J.A.E, Hytonen, V.P, Airenne, T.T, Niskanen, E, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-02-14 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Modification of Ligand-Binding Preference of Avidin by Circular Permutation and Mutagenesis.
Chembiochem, 9, 2008
|
|
5KWV
 
 | |
1GUS
 
 | |
4A85
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with kinetin. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
1GUT
 
 | |
1PXK
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
3E5K
 
 | | Crystal structure of CYP105P1 wild-type 4-phenylimidazole complex | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
1PXL
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-(4-trifluoromethyl-phenyl)-amine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
4H2T
 
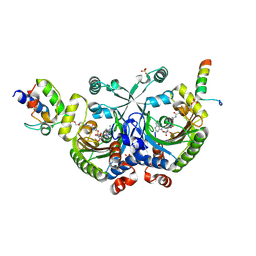 | | Crystal structure of Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with cognate carrier protein and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
3M62
 
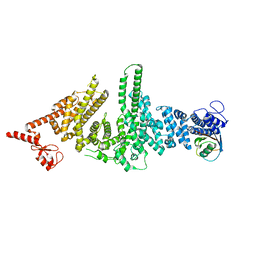 | |
1S8I
 
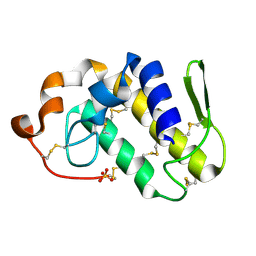 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, second fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
6O14
 
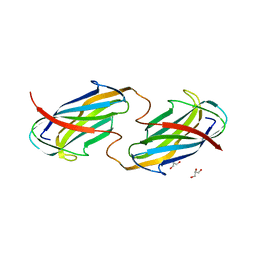 | |
3HM1
 
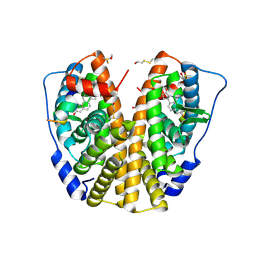 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren-17-one) | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Liwanag, M, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one)
To be Published
|
|
4PP0
 
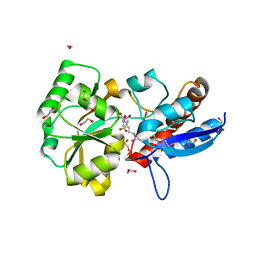 | | Structure of the PBP NocT-M117N in complex with pyronopaline | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
4A83
 
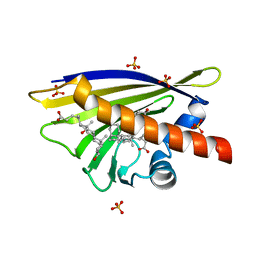 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with deoxycholate. | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
3E5L
 
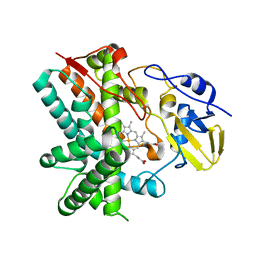 | | Crystal structure of CYP105P1 H72A mutant | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
8IJB
 
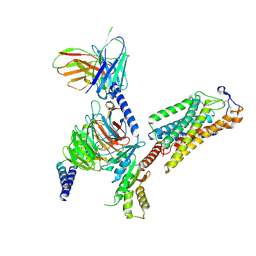 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
3GER
 
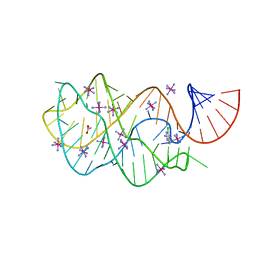 | | Guanine riboswitch bound to 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Batey, R.T. | | Deposit date: | 2009-02-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
4AP2
 
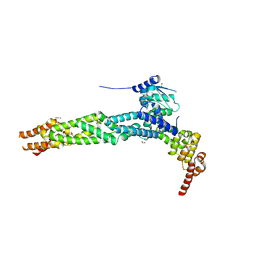 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
2XG3
 
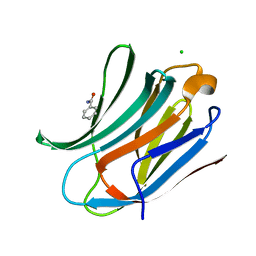 | | Human galectin-3 in complex with a benzamido-N-acetyllactoseamine inhibitor | | Descriptor: | BENZAMIDE, CHLORIDE ION, Galectin-3, ... | | Authors: | Diehl, C, Engstrom, O, Delaine, T, Hakansson, M, Genheden, S, Modig, K, Leffler, H, Ryde, U, Nilsson, U, Akke, M. | | Deposit date: | 2010-05-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J. Am. Chem. Soc., 132, 2010
|
|
2EVS
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
4OXR
 
 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | Descriptor: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
1FQD
 
 | | CRYSTAL STRUCTURE OF MALTOTETRAITOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
